From expression footprints to causal pathways: contextualizing large signaling networks with CARNIVAL
- PMID: 31728204
- PMCID: PMC6848167
- DOI: 10.1038/s41540-019-0118-z
From expression footprints to causal pathways: contextualizing large signaling networks with CARNIVAL
Abstract
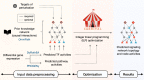
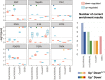
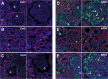
While gene expression profiling is commonly used to gain an overview of cellular processes, the identification of upstream processes that drive expression changes remains a challenge. To address this issue, we introduce CARNIVAL, a causal network contextualization tool which derives network architectures from gene expression footprints. CARNIVAL (CAusal Reasoning pipeline for Network identification using Integer VALue programming) integrates different sources of prior knowledge including signed and directed protein-protein interactions, transcription factor targets, and pathway signatures. The use of prior knowledge in CARNIVAL enables capturing a broad set of upstream cellular processes and regulators, leading to a higher accuracy when benchmarked against related tools. Implementation as an integer linear programming (ILP) problem guarantees efficient computation. As a case study, we applied CARNIVAL to contextualize signaling networks from gene expression data in IgA nephropathy (IgAN), a condition that can lead to chronic kidney disease. CARNIVAL identified specific signaling pathways and associated mediators dysregulated in IgAN including Wnt and TGF-β, which we subsequently validated experimentally. These results demonstrated how CARNIVAL generates hypotheses on potential upstream alterations that propagate through signaling networks, providing insights into diseases.
Keywords: Regulatory networks; Software.
© The Author(s) 2019.
Conflict of interest statement
Competing interestsThe authors declare no competing interests
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

