RbfA and IF3 couple ribosome biogenesis and translation initiation to increase stress tolerance
- PMID: 31728529
- PMCID: PMC7145577
- DOI: 10.1093/nar/gkz1065
RbfA and IF3 couple ribosome biogenesis and translation initiation to increase stress tolerance
Abstract
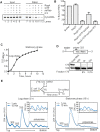
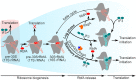
Bacterial ribosome biogenesis and translation occur in the same cellular compartment. Therefore, a biochemical gate-keeping step is required to prevent error-prone immature ribosomes from engaging in protein synthesis. Here, we provide evidence for a previously unknown quality control mechanism in which the abundant ribosome assembly factor, RbfA, suppresses protein synthesis by immature Escherichia coli 30S subunits. After 30S maturation, RbfA is displaced by initiation factor 3 (IF3), which promotes translation initiation. Genetic interactions between RbfA and IF3 show that RbfA release by IF3 is important during logarithmic growth as well as during stress encountered during stationary phase, low nutrition, low temperature, and antibiotics. By gating the transition from 30S biogenesis to translation initiation, RbfA and IF3 maintain the fidelity of bacterial protein synthesis.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
Era and RbfA have overlapping function in ribosome biogenesis in Escherichia coli.J Mol Microbiol Biotechnol. 2006;11(1-2):41-52. doi: 10.1159/000092818. J Mol Microbiol Biotechnol. 2006. PMID: 16825789
-
Cold-stress-induced de novo expression of infC and role of IF3 in cold-shock translational bias.RNA. 2007 Aug;13(8):1355-65. doi: 10.1261/rna.455607. Epub 2007 Jun 25. RNA. 2007. PMID: 17592046 Free PMC article.
-
The role of RbfA in 16S rRNA processing and cell growth at low temperature in Escherichia coli.J Mol Biol. 2003 Sep 19;332(3):575-84. doi: 10.1016/s0022-2836(03)00953-7. J Mol Biol. 2003. PMID: 12963368
-
Mechanism of recycling of post-termination ribosomal complexes in eubacteria: a new role of initiation factor 3.J Biosci. 2006 Jun;31(2):281-9. doi: 10.1007/BF02703921. J Biosci. 2006. PMID: 16809861 Review.
-
Assembly of bacterial ribosomes.Annu Rev Biochem. 2011;80:501-26. doi: 10.1146/annurev-biochem-062608-160432. Annu Rev Biochem. 2011. PMID: 21529161 Review.
Cited by
-
Protein Assistants of Small Ribosomal Subunit Biogenesis in Bacteria.Microorganisms. 2022 Mar 30;10(4):747. doi: 10.3390/microorganisms10040747. Microorganisms. 2022. PMID: 35456798 Free PMC article. Review.
-
Differential Proteomic Analysis of Listeria monocytogenes during High-Pressure Processing.Biology (Basel). 2022 Jul 31;11(8):1152. doi: 10.3390/biology11081152. Biology (Basel). 2022. PMID: 36009779 Free PMC article.
-
Nucleolar origins: challenging perspectives on evolution and function.Open Biol. 2025 Mar;15(3):240330. doi: 10.1098/rsob.240330. Epub 2025 Mar 12. Open Biol. 2025. PMID: 40068812 Free PMC article. Review.
-
A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.Sci Adv. 2021 Jun 4;7(23):eabf7547. doi: 10.1126/sciadv.abf7547. Print 2021 Jun. Sci Adv. 2021. PMID: 34088665 Free PMC article.
-
Epitranscriptomics of Mammalian Mitochondrial Ribosomal RNA.Cells. 2020 Sep 27;9(10):2181. doi: 10.3390/cells9102181. Cells. 2020. PMID: 32992603 Free PMC article. Review.
References
-
- Shajani Z., Sykes M.T., Williamson J.R.. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 2011; 80:501–526. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

