Mechanisms regulating PD-L1 expression on tumor and immune cells
- PMID: 31730010
- PMCID: PMC6858680
- DOI: 10.1186/s40425-019-0770-2
Mechanisms regulating PD-L1 expression on tumor and immune cells
Abstract
Background: The PD-1/PD-L1 checkpoint is a central mediator of immunosuppression in the tumor immune microenvironment (TME) and is primarily associated with IFN-g signaling. To characterize other factors regulating PD-L1 expression on tumor and/or immune cells, we investigated TME-resident cytokines and the role of transcription factors in constitutive and cytokine-induced PD-L1 expression.
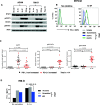
Methods: Thirty-four cultured human tumor lines [18 melanomas (MEL), 12 renal cell carcinomas (RCC), 3 squamous cell carcinomas of the head and neck (SCCHN), and 1 non-small-cell lung carcinoma (NSCLC)] and peripheral blood monocytes (Monos) were treated with cytokines that we detected in the PD-L1+ TME by gene expression profiling, including IFN-g, IL-1a, IL-10, IL-27 and IL-32g. PD-L1 cell surface protein expression was detected by flow cytometry, and mRNA by quantitative real-time PCR. Total and phosphorylated STAT1, STAT3, and p65 proteins were detected by Western blotting, and the genes encoding these proteins were knocked down with siRNAs. Additionally, the proximal promoter region of PDL1 (CD274) was sequenced in 33 cultured tumors.
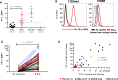
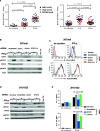
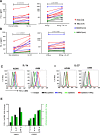
Results: PD-L1 was constitutively expressed on 1/17 cultured MELs, 8/11 RCCs, 3/3 SCCHNs, and on Monos. Brief IFN-g exposure rapidly induced PD-L1 on all tumor cell lines and Monos regardless of constitutive PD-L1 expression. PD-L1 mRNA levels were associated with protein expression, which was diminished by exposure to transcriptional inhibitors. siRNA knockdown of STAT1 but not STAT3 reduced IFN-g- and IL-27-induced PD-L1 protein expression on tumor cells. In contrast, STAT3 knockdown in Monos reduced IL-10-induced PD-L1 protein expression, and p65 knockdown in tumor cells reduced IL-1a-induced PD-L1 expression. Notably, constitutive PD-L1 expression was not affected by knocking down STAT1, STAT3, or p65. Differential effects of IFN-g, IL-1a, and IL-27 on individual tumor cell lines were not due to PDL1 promoter polymorphisms.
Conclusions: Multiple cytokines found in an immune-reactive TME may induce PD-L1 expression on tumor and/or immune cells through distinct signaling mechanisms. Factors driving constitutive PD-L1 expression were not identified in this study. Understanding complex mechanisms underlying PD-L1 display in the TME may allow treatment approaches mitigating expression of this immunosuppressive ligand, to enhance the impact of PD-1 blockade.
Keywords: Cancer immunotherapy; Cytokines; Interferon gamma; Interleukins; PD-L1; Transcription factors; Tumor microenvironment.
Conflict of interest statement
DMP and SLT report stock and other ownership interests from Aduro Biotech, Compugen, DNAtrix, Dragonfly Therapeutics, ERVAXX, Five Prime Therapeutics, FLX Bio, Jounce Therapeutics, Potenza Therapeutics, Tizona Therapeutics, and WindMIL; consulting or advisory role with AbbVie, Amgen, Bayer, Compugen, DNAtrix, Dragonfly Therapeutics, Dynavax, ERVAXX, Five Prime Therapeutics, FLX Bio, lmmunomic Therapeutics, Janssen Oncology, Medlmmune, Merck, Tizona Therapeutics, and Wind MIL; research funding from Bristol-Myers Squibb, Compugen, and Potenza Therapeutics; patents, royalties, and other intellectual property from Aduro Biotech, Bristol-Myers Squibb, and lmmunonomic Therapeutics; and travel, accommodations, expenses from Bristol-Myers Squibb, and Five Prime Therapeutics. SC, GAC, TSP, PW, TLM and FP have no conflicts of interest to disclose.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
