A Novel cis Element Achieves the Same Solution as an Ancestral cis Element During Thiamine Starvation in Candida glabrata
- PMID: 31732505
- PMCID: PMC6945020
- DOI: 10.1534/g3.119.400897
A Novel cis Element Achieves the Same Solution as an Ancestral cis Element During Thiamine Starvation in Candida glabrata
Abstract
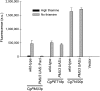
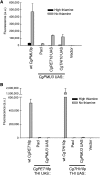
Regulatory networks often converge on very similar cis sequences to drive transcriptional programs due to constraints on what transcription factors are present. To determine the role of constraint loss on cis element evolution, we examined the recent appearance of a thiamine starvation regulated promoter in Candida glabrata This species lacks the ancestral transcription factor Thi2, but still has the transcription factor Pdc2, which regulates thiamine starvation genes, allowing us to determine the effect of constraint change on a new promoter. We identified two different cis elements in C. glabrata - one present in the evolutionarily recent gene called CgPMU3, and the other element present in the other thiamine (THI) regulated genes. Reciprocal swaps of the cis elements and incorporation of the S. cerevisiae Thi2 transcription factor-binding site into these promoters demonstrate that the two elements are functionally different from one another. Thus, this loss of an imposed constraint on promoter function has generated a novel cis sequence, suggesting that loss of trans constraints can generate a non-convergent pathway with the same output.
Keywords: Candida glabrata; PDC2; THI2; cis evolution; thiamine.
Copyright © 2020 Iosue et al.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
