Assessing Protein Sequence Database Suitability Using De Novo Sequencing
- PMID: 31732549
- PMCID: PMC6944239
- DOI: 10.1074/mcp.TIR119.001752
Assessing Protein Sequence Database Suitability Using De Novo Sequencing
Abstract
The analysis of samples from unsequenced and/or understudied species as well as samples where the proteome is derived from multiple organisms poses two key questions. The first is whether the proteomic data obtained from an unusual sample type even contains peptide tandem mass spectra. The second question is whether an appropriate protein sequence database is available for proteomic searches. We describe the use of automated de novo sequencing for evaluating both the quality of a collection of tandem mass spectra and the suitability of a given protein sequence database for searching that data. Applications of this method include the proteome analysis of closely related species, metaproteomics, and proteomics of extinct organisms.
Keywords: Algorithms; Caenorhabditis elegans; data evaluation; de novo sequencing; mass spectrometry; metaproteomics; peptides*; protein identification; quality control and metrics; sequencing ms; tandem mass spectrometry.
© 2020 Johnson et al.
Figures


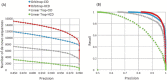

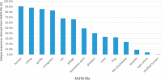

References
-
- Cilia M., Tamborindeguy C., Rolland M., Howe K., Thannhauser T. W., and Gray S. (2011) Tangible benefits of the aphid Acyrthosiphon pisum genome sequencing for aphid proteomics: Enhancements in protein identification and data validation for homology-based proteomics. J. Insect Physiol. 57, 179–190 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

