Prion disease diagnosis using subject-specific imaging biomarkers within a multi-kernel Gaussian process
- PMID: 31734530
- PMCID: PMC6978211
- DOI: 10.1016/j.nicl.2019.102051
Prion disease diagnosis using subject-specific imaging biomarkers within a multi-kernel Gaussian process
Abstract
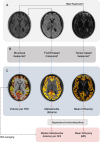
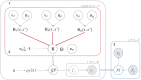
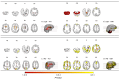
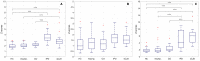
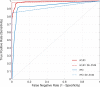
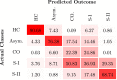
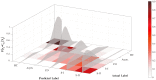
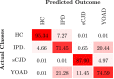
Prion diseases are a group of rare neurodegenerative conditions characterised by a high rate of progression and highly heterogeneous phenotypes. Whilst the most common form of prion disease occurs sporadically (sporadic Creutzfeldt-Jakob disease, sCJD), other forms are caused by prion protein gene mutations, or exposure to prions in the diet or by medical procedures, such us surgeries. To date, there are no accurate quantitative imaging biomarkers that can be used to predict the future clinical diagnosis of a healthy subject, or to quantify the progression of symptoms over time. Besides, CJD is commonly mistaken for other forms of dementia. Due to the heterogeneity of phenotypes and the lack of a consistent geometrical pattern of disease progression, the approaches used to study other types of neurodegenerative diseases are not satisfactory to capture the progression of human form of prion disease. In this paper, using a tailored framework, we aim to classify and stratify patients with prion disease, according to the severity of their illness. The framework is initialised with the extraction of subject-specific imaging biomarkers. The extracted biomakers are then combined with genetic and demographic information within a Gaussian Process classifier, used to calculate the probability of a subject to be diagnosed with prion disease in the next year. We evaluate the effectiveness of the proposed method in a cohort of patients with inherited and sporadic forms of prion disease. The model has shown to be effective in the prediction of both inherited CJD (92% of accuracy) and sporadic CJD (95% of accuracy). However the model has shown to be less effective when used to stratify the different stages of the disease, in which the average accuracy is 85%, whilst the recall is 59%. Finally, our framework was extended as a differential diagnosis tool to identify both forms of CJD among another neurodegenerative disease. In summary we have developed a novel method for prion disease diagnosis and prediction of clinical onset using multiple sources of features, which may have use in other disorders with heterogeneous imaging features.
Keywords: Biomarkers; Diagnosis; Gaussian process; Inherited Creutzfeldt–Jakob disease; Prion diseases; Sporadic Creutzfeldt–Jakob disease; Subjects’ stratification.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
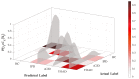
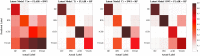
References
-
- Alner K., Hyare H., Mead S., Rudge P., Wroe S., Rohrer J.D., Ridgway G.R., Ourselin S., Clarkson M., Hunt H., Fox N.C., Webb T., Collinge J., Cipolotti L. Distinct neuropsychological profiles correspond to distribution of cortical thinning in inherited prion disease caused by insertional mutation. J. Neurol. Neurosurg. Psychiatr. 2012;83(1):109–114. - PubMed
-
- Bradford B.M., Piccardo P., Ironside J.W., Mabbott N.A. Human prion diseases and the risk of their transmission during anatomical dissection. Clinical Anatomy. 2014;27(6):821–832. - PubMed
-
- Canas L.S., Yvernault B., Cash D.M., Molteni E., Veale T., Benzinger T., Ourselin S., Mead S., Modat M. Gaussian processes with optimal kernel construction for neuro-degenerative clinical onset prediction. Houston. 2018;10575:10575–10576.
-
- Canas L.S., Yvernault B., Sudre C., Vita E.D., Cardoso M.J., Canas L.S., Yvernault B., Sudre C., Vita E.D., Cardoso J., Thornton J., Barkhof F., Ourselin S., Mead S., Modat M. Proc. SPIE 10574, Medical Imaging 2018: Imaging Processing. 1057405. 2018. Imaging biomarkers for the diagnosis of Prion disease.
-
- Caobelli F., Cobelli M., Pizzocaro C., Pavia M., Magnaldi S., Guerra U.P. The role of neuroimaging in evaluating patients affected by Creutzfeldt–Jakob disease: A Systematic review of the literature. J. Neuroimaging. 2014;6:1–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

