YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma
- PMID: 31735169
- PMCID: PMC6859620
- DOI: 10.1186/s12943-019-1082-3
YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma
Erratum in
-
Correction to: YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma.Mol Cancer. 2020 Sep 4;19(1):137. doi: 10.1186/s12943-020-01257-8. Mol Cancer. 2020. PMID: 32887624 Free PMC article.
Abstract
Background: Dynamic N6-methyladenosine (m6A) modification was previously identified as a ubiquitous post-transcriptional regulation that affected mRNA homeostasis. However, the m6A-related epitranscriptomic alterations and functions remain elusive in human cancer. Here we aim to identify the profile and outcome of m6A-methylation in hepatocellular carcinoma (HCC).
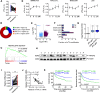
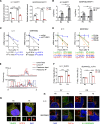
Results: Using liquid chromatography-tandem mass spectrometry and m6A-immunoprecipitation in combination with high-throughput sequencing, we determined the m6A-mRNA levels in human HCC. Human HCC exhibited a characteristic gain of m6A modification in tandem with an increase of mRNA expression, owing to YTH domain family 2 (YTHDF2) reduction. The latter predicted poor classification and prognosis of HCC patients, and highly correlated with HCC m6A landscape. YTHDF2 silenced in human HCC cells or ablated in mouse hepatocytes provoked inflammation, vascular reconstruction and metastatic progression. Mechanistically, YTHDF2 processed the decay of m6A-containing interleukin 11 (IL11) and serpin family E member 2 (SERPINE2) mRNAs, which were responsible for the inflammation-mediated malignancy and disruption of vascular normalization. Reciprocally, YTHDF2 transcription succumbed to hypoxia-inducible factor-2α (HIF-2α). Administration of a HIF-2α antagonist (PT2385) restored YTHDF2-programed epigenetic machinery and repressed liver cancer.
Conclusion: Our results have characterized the m6A-mRNA landscape in human HCC and revealed YTHDF2 as a molecular 'rheostat' in epitranscriptome and cancer progression.
Keywords: HCC; HIF-2α antagonism; IL-11; Inflammation; Serpin E2; Vessel normalization; YTHDF2; m6A.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

