Selective Persulfide Detection Reveals Evolutionarily Conserved Antiaging Effects of S-Sulfhydration
- PMID: 31735592
- PMCID: PMC7185476
- DOI: 10.1016/j.cmet.2019.10.007
Selective Persulfide Detection Reveals Evolutionarily Conserved Antiaging Effects of S-Sulfhydration
Erratum in
-
Selective Persulfide Detection Reveals Evolutionarily Conserved Antiaging Effects of S-Sulfhydration.Cell Metab. 2020 Jan 7;31(1):207. doi: 10.1016/j.cmet.2019.12.001. Cell Metab. 2020. PMID: 31914376 Free PMC article. No abstract available.
Abstract
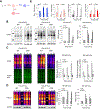
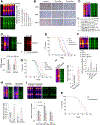
Life on Earth emerged in a hydrogen sulfide (H2S)-rich environment eons ago and with it protein persulfidation mediated by H2S evolved as a signaling mechanism. Protein persulfidation (S-sulfhydration) is a post-translational modification of reactive cysteine residues, which modulate protein structure and/or function. Persulfides are difficult to label and study due to their reactivity and similarity with cysteine. Here, we report a facile strategy for chemoselective persulfide bioconjugation using dimedone-based probes, to achieve highly selective, rapid, and robust persulfide labeling in biological samples with broad utility. Using this method, we show persulfidation is an evolutionarily conserved modification and waves of persulfidation are employed by cells to resolve sulfenylation and prevent irreversible cysteine overoxidation preserving protein function. We report an age-associated decline in persulfidation that is conserved across evolutionary boundaries. Accordingly, dietary or pharmacological interventions to increase persulfidation associate with increased longevity and improved capacity to cope with stress stimuli.
Keywords: aging; calorie restriction; hydrogen peroxide; hydrogen sulfide; protein persulfidation; redox signaling; sulfenylation; sulfinylation; sulfonylation.
Crown Copyright © 2019. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures





Comment in
-
A Central Role for the Gasotransmitter H2S in Aging.Cell Metab. 2020 Jan 7;31(1):10-12. doi: 10.1016/j.cmet.2019.11.015. Epub 2019 Dec 5. Cell Metab. 2020. PMID: 31951564
References
-
- Alexander BE, Coles SJ, Fox BC, Khan TF, Maliszewski J, Perry A, Pitak MB, Whiteman M, Wood ME, Nakashima I, et al. (2015). Investigating the generation of hydrogen sulfide from the phosphonamidodithioate slow-release donor GYY4137. Med. Chem. Commun 6, 1649–1655.
-
- Artaud I, and Galardon E (2014). A persulfide analogue of the nitrosothiol SNAP: formation, characterization and reactivity. Chembiochem 15, 2361–2364. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

