Mutations in ANKLE2, a ZIKA Virus Target, Disrupt an Asymmetric Cell Division Pathway in Drosophila Neuroblasts to Cause Microcephaly
- PMID: 31735666
- PMCID: PMC6917859
- DOI: 10.1016/j.devcel.2019.10.009
Mutations in ANKLE2, a ZIKA Virus Target, Disrupt an Asymmetric Cell Division Pathway in Drosophila Neuroblasts to Cause Microcephaly
Abstract
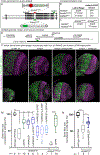
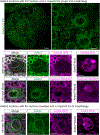
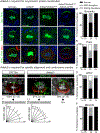
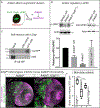
The apical Par complex, which contains atypical protein kinase C (aPKC), Bazooka (Par-3), and Par-6, is required for establishing polarity during asymmetric division of neuroblasts in Drosophila, and its activity depends on L(2)gl. We show that loss of Ankle2, a protein associated with microcephaly in humans and known to interact with Zika protein NS4A, reduces brain volume in flies and impacts the function of the Par complex. Reducing Ankle2 levels disrupts endoplasmic reticulum (ER) and nuclear envelope morphology, releasing the kinase Ballchen-VRK1 into the cytosol. These defects are associated with reduced phosphorylation of aPKC, disruption of Par-complex localization, and spindle alignment defects. Importantly, removal of one copy of ballchen or l(2)gl suppresses Ankle2 mutant phenotypes and restores viability and brain size. Human mutational studies implicate the above-mentioned genes in microcephaly and motor neuron disease. We suggest that NS4A, ANKLE2, VRK1, and LLGL1 define a pathway impinging on asymmetric determinants of neural stem cell division.
Keywords: Ballchen; Bazooka; L(2)gl; MCPH16; Miranda; NS4A; VRK1; aPKC; brain development; congenital infection.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures


References
-
- Asencio C, Davidson IF, Santarella-Mellwig R, Ly-Hartig TB, Mall M, Wallenfang MR, Mattaj IW and Gorjanacz M (2012) ‘Coordination of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis’, Cell, 150(1), pp. 122–35. - PubMed

