Temporal Dynamics of Co-circulating Lineages of Porcine Reproductive and Respiratory Syndrome Virus
- PMID: 31736919
- PMCID: PMC6839445
- DOI: 10.3389/fmicb.2019.02486
Temporal Dynamics of Co-circulating Lineages of Porcine Reproductive and Respiratory Syndrome Virus
Abstract
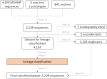
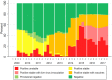
Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) is the most important endemic pathogen in the U.S. swine industry. Despite control efforts involving improved biosecurity and different vaccination protocols, the virus continues to circulate and evolve. One of the foremost challenges in its control is high levels of genetic and antigenic diversity. Here, we quantify the co-circulation, emergence and sequential turnover of multiple PRRSV lineages in a single swine-producing region in the United States over a span of 9 years (2009-2017). By classifying over 4,000 PRRSV sequences (open-reading frame 5) into phylogenetic lineages and sub-lineages, we document the ongoing diversification and temporal dynamics of the PRRSV population, including the rapid emergence of a novel sub-lineage that appeared to be absent globally pre-2008. In addition, lineage 9 was the most prevalent lineage from 2009 to 2010, but its occurrence fell to 0.5% of all sequences identified per year after 2014, coinciding with the emergence or re-emergence of lineage 1 as the dominant lineage. The sequential dominance of different lineages, as well as three different sub-lineages within lineage 1, is consistent with the immune-mediated selection hypothesis for the sequential turnover in the dominant lineage. As host populations build immunity through natural infection or vaccination toward the most common variant, this dominant (sub-) lineage may be replaced by an emerging variant to which the population is more susceptible. An analysis of patterns of non- synonymous and synonymous mutations revealed evidence of positive selection on immunologically important regions of the genome, further supporting the potential that immune-mediated selection shapes the evolutionary and epidemiological dynamics for this virus. This has important implications for patterns of emergence and re-emergence of genetic variants of PRRSV that have negative impacts on the swine industry. Constant surveillance on PRRSV occurrence is crucial to a better understanding of the epidemiological and evolutionary dynamics of co-circulating viral lineages. Further studies utilizing whole genome sequencing and exploring the extent of cross-immunity between heterologous PRRS viruses could shed further light on PRRSV immunological response and aid in developing strategies that might be able to diminish disease impact.
Keywords: PRRSV; ecology; emergence; epidemiology; evolution; multi-strain dynamics; outbreak.
Copyright © 2019 Paploski, Corzo, Rovira, Murtaugh, Sanhueza, Vilalta, Schroeder and VanderWaal.
Figures




References
-
- Animal Disease Research and Diagnostic Laboratory, and South Dakota State Univeristy (2017). DNA Sequencing of PRRSV using ORF 5 PCR - MOL.SOP.0007.08. Brookings, SD: South Dakota State Univeristy.
-
- Ansari I. H., Kwon B., Osorio F. A., Pattnaik A. K. (2006). Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J. Virol. 80 3994–4004. 10.1128/jvi.80.8.3994-4004.2006 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

