MiR-155 Regulates PAD4-Dependent Formation of Neutrophil Extracellular Traps
- PMID: 31736940
- PMCID: PMC6838784
- DOI: 10.3389/fimmu.2019.02462
MiR-155 Regulates PAD4-Dependent Formation of Neutrophil Extracellular Traps
Abstract
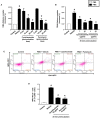
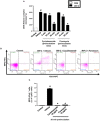
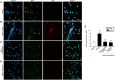
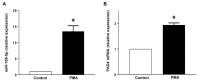
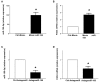
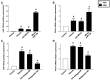
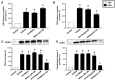
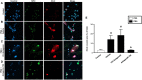
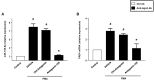
Accumulating data suggest that neutrophil extracellular traps (NETs) exert a key function in several diseases. Peptidylarginine deiminase 4 (PAD4) regulates NET formation via citrullination of histones. The aim of this study was to examine the role of miR-155 in controlling PAD4-dependent generation of NETs. Bone marrow neutrophils were stimulated with PMA and MIP-2. Pre-incubation of neutrophils with translational inhibitors (cycloheximide or puromycin) markedly decreased NET formation induced by PMA or MIP-2. Neutrophil transfection with a mimic miR-155 increased PMA-induced PAD4 mRNA expression and NET formation. In contrast, transfection with an antagomiR-155 decreased induction of PAD4 mRNA and NETs in response to PMA challenge. Bioinformatical examination of PAD4 revealed a potential binding site in AU-rich elements at the 3'-UTR region. MiR-155 binding to PAD4 was examined by use of target site blockers and RNA immunoprecipitation, revealing that miR-155 regulation of PAD4 mRNA is mediated via AU-rich elements in the 3'-UTR region. In conclusion, our findings demonstrate that miR-155 positively regulates neutrophil expression of PAD4 and expulsion of extracellular traps. Thus, our novel results indicate that targeting miR-155 might be useful to inhibit exaggerated NET generation in inflammatory diseases.
Keywords: NETosis; histone; inflammation; microRNA; neutrophils.
Copyright © 2019 Hawez, Al-Haidari, Madhi, Rahman and Thorlacius.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

