Improving Short- and Long-Term Genetic Gain by Accounting for Within-Family Variance in Optimal Cross-Selection
- PMID: 31737033
- PMCID: PMC6828944
- DOI: 10.3389/fgene.2019.01006
Improving Short- and Long-Term Genetic Gain by Accounting for Within-Family Variance in Optimal Cross-Selection
Abstract
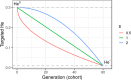
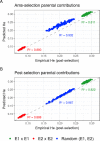
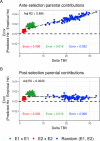
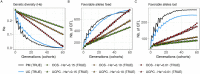
The implementation of genomic selection in recurrent breeding programs raises the concern that a higher inbreeding rate could compromise the long-term genetic gain. An optimized mating strategy that maximizes the performance in progeny and maintains diversity for long-term genetic gain is therefore essential. The optimal cross-selection approach aims at identifying the optimal set of crosses that maximizes the expected genetic value in the progeny under a constraint on genetic diversity in the progeny. Optimal cross-selection usually does not account for within-family selection, i.e., the fact that only a selected fraction of each family is used as parents of the next generation. In this study, we consider within-family variance accounting for linkage disequilibrium between quantitative trait loci to predict the expected mean performance and the expected genetic diversity in the selected progeny of a set of crosses. These predictions rely on the usefulness criterion parental contribution (UCPC) method. We compared UCPC-based optimal cross-selection and the optimal cross-selection approach in a long-term simulated recurrent genomic selection breeding program considering overlapping generations. UCPC-based optimal cross-selection proved to be more efficient to convert the genetic diversity into short- and long-term genetic gains than optimal cross-selection. We also showed that, using the UCPC-based optimal cross-selection, the long-term genetic gain can be increased with only a limited reduction of the short-term commercial genetic gain.
Keywords: Bulmer effect; genetic diversity; genomic prediction; optimal cross-selection; parental contributions; usefulness criterion.
Copyright © 2019 Allier, Lehermeier, Charcosset, Moreau and Teyssèdre.
Figures




References
-
- Allier A., Teyssèdre S., Lehermeier C., Claustres B., Maltese S., Moreau L., Charcosset A. (2019. a). Assessment of breeding programs sustainability: application of phenotypic and genomic indicators to a North European grain maize program. Theor. Appl. Genet. 132, 1321–1334. 10.1007/s00122-019-03280-w - DOI - PubMed
-
- Bernardo R., Moreau L., Charcosset A. (2006). Number and fitness of selected individuals in marker-assisted and phenotypic recurrent selection. Crop Sci. 46, 1972–1980. 10.2135/cropsci2006.01-0057 - DOI
LinkOut - more resources
Full Text Sources

