Perspectives on miRNAs as Epigenetic Markers in Osteoporosis and Bone Fracture Risk: A Step Forward in Personalized Diagnosis
- PMID: 31737038
- PMCID: PMC6831724
- DOI: 10.3389/fgene.2019.01044
Perspectives on miRNAs as Epigenetic Markers in Osteoporosis and Bone Fracture Risk: A Step Forward in Personalized Diagnosis
Abstract
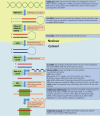
Aging is associated with an increased incidence of age-related bone diseases. Current diagnostics (e.g., conventional radiology, biochemical markers), because limited in specificity and sensitivity, can distinguish between healthy or osteoporotic subjects but they are unable to discriminate among different underlying causes that lead to the same bone pathological condition (e.g., bone fracture risk). Among recent, more sensitive biomarkers, miRNAs - the non-coding RNAs involved in the epigenetic regulation of gene expression, have emerged as fundamental post-transcriptional modulators of bone development and homeostasis. Each identified miRNA carries out a specific role in osteoblast and osteoclast differentiation and functional pathways (osteomiRs). miRNAs bound to proteins or encapsulated in exosomes and/or microvesicles are released into the bloodstream and biological fluids where they can be detected and measured by highly sensitive and specific methods (e.g., quantitative PCR, next-generation sequencing). As such, miRNAs provide a prompt and easily accessible tool to determine the subject-specific epigenetic environment of a specific condition. Their use as biomarkers opens new frontiers in personalized medicine. While miRNAs circulating levels are lower than those found in the tissue/cell source, their quantification in biological fluids may be strategic in the diagnosis of diseases that affect tissues, such as bone, in which biopsy may be especially challenging. For a biomarker to be valuable in clinical practice and support medical decisions, it must be (easily) measurable, validated by independent studies, and strongly and significantly associated with a disease outcome. Currently, miRNAs analysis does not completely satisfy these criteria, however. Starting from in vitro and in vivo observations describing their biological role in bone cell development and metabolism, this review describes the potential use of bone-associated circulating miRNAs as biomarkers for determining predisposition, onset, and development of osteoporosis and bone fracture risk. Moreover, the review focuses on their clinical relevance and discusses the pre-analytical, analytical, and post-analytical issues in their measurement, which still limits their routine application. Taken together, research and clinical findings may be helpful for creating miRNA-based diagnostic tools in the diagnosis and treatment of bone diseases.
Keywords: biomarkers; circulating miRNAs; extra-analytical variability; fracture risk; miRNA signature; osteopenia/osteoporosis; sensitivity and specificity.
Copyright © 2019 Bottani, Banfi and Lombardi.
Figures
References
-
- Andersen C. L., Jensen J. L., Orntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Re.s 64 (15), 5245–5250. 10.1158/0008-5472.CAN-04-0496 - DOI - PubMed
-
- Arroyo J. D., Chevillet J. R., Kroh E. M., Ruf I. K., Pritchard C. C., Gibson D. F., et al. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U. S. A. 108 (12), 5003–5008. 10.1073/pnas.1019055108 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

