Genotyping and Phylogenetic Position of Trichinella spiralis Isolates from Different Geographical Locations in China
- PMID: 31737057
- PMCID: PMC6834790
- DOI: 10.3389/fgene.2019.01093
Genotyping and Phylogenetic Position of Trichinella spiralis Isolates from Different Geographical Locations in China
Abstract
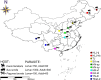
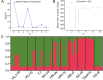
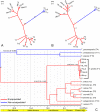
In China, the nematode Trichinella spiralis is the main aetiological agent of human trichinellosis. We performed multi-locus microsatellite typing of T. spiralis isolates to improve the current knowledge of the evolution and population diversity. First, seven polymorphic microsatellite loci were used to infer the genetic diversity of T. spiralis collected in 10 endemic regions. Then, a Bayesian model-based STRUCTURE analysis, a clustering based on the neighbor-joining method, and a principal coordinate analysis (PCA) were performed to identify the genetic structure. Finally, the phylogenetic position of Chinese isolates was explored based on six mitochondrial and nuclear genetic markers (cox1, cytb, 5S ISR, ESV, ITS1, and 18S rDNA) using the maximum likelihood and Bayesian methods. In addition, the divergence time was estimated with multiple genes using an uncorrelated log-normal relaxed molecular-clock model. A total of 16 alleles were detected in 2,310 individuals (1,650 muscle larvae and 660 adult worms) using seven loci. The STRUCTURE analysis indicated that the T. spiralis isolates could be organized and derived from the admixture of two ancestral clusters, which was also substantiated through the clustering analysis based on the allelic data. PCA separated most samples from Tiandong, Guangxi (GX-td), and Linzhi, Tibet (Tibet-lz), from the remaining isolates. However, both maximum likelihood and Bayesian inference supported the close relationship between Xiangfan, Hubei (HB-xf), and GX-td. The molecular dating analysis suggested that the Chinese isolates started to diverge during the Late Pleistocene (0.69 Mya). Generally, T. spiralis was observed to harbor low genetic variation, and further investigation with deeper sampling is needed to elucidate the population structure.
Keywords: China; Trichinella spiralis; genetic variance; microsatellite; population.
Copyright © 2019 Zhang, Han, Hong, Jiang, Niu, Wang and Cui.
Figures


References
-
- Aggarwal R. K., Hendre P. S., Varshney R. K., Bhat P. R., Krishnakumar V., Singh L. (2007). Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theor. Appl. Genet. 114, 359–372. 10.1007/s00122-006-0440-x - DOI - PubMed
-
- Earl D. A., von Holdt B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. 10.1007/s12686-011-9548-7 - DOI
LinkOut - more resources
Full Text Sources

