A combined structural and biochemical approach reveals translocation and stalling of UvrB on the DNA lesion as a mechanism of damage verification in bacterial nucleotide excision repair
- PMID: 31739207
- PMCID: PMC7616783
- DOI: 10.1016/j.dnarep.2019.102746
A combined structural and biochemical approach reveals translocation and stalling of UvrB on the DNA lesion as a mechanism of damage verification in bacterial nucleotide excision repair
Abstract
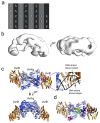
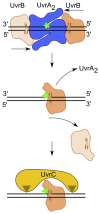
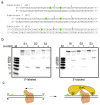
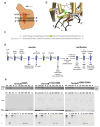
Nucleotide excision repair (NER) is a DNA repair pathway present in all domains of life. In bacteria, UvrA protein localizes the DNA lesion, followed by verification by UvrB helicase and excision by UvrC double nuclease. UvrA senses deformations and flexibility of the DNA duplex without precisely localizing the lesion in the damaged strand, an element essential for proper NER. Using a combination of techniques, we elucidate the mechanism of the damage verification step in bacterial NER. UvrA dimer recruits two UvrB molecules to its two sides. Each of the two UvrB molecules clamps a different DNA strand using its β-hairpin element. Both UvrB molecules then translocate to the lesion, and UvrA dissociates. The UvrB molecule that clamps the damaged strand gets stalled at the lesion to recruit UvrC. This mechanism allows UvrB to verify the DNA damage and identify its precise location triggering subsequent steps in the NER pathway.
Keywords: DNA repair; Prokaryotic nucleotide excision repair; UvrA; UvrB; UvrC.
Copyright © 2019 The Authors. Published by Elsevier B.V. All rights reserved.
Figures

References
-
- Sancar A, Reardon JT. Nucleotide excision repair in E. Coli and man. Adv Protein Chem. 2004;69:43–71. - PubMed
-
- Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem Rev. 2006;106:233–252. - PubMed
-
- Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. - PubMed

