RepeatFiller newly identifies megabases of aligning repetitive sequences and improves annotations of conserved non-exonic elements
- PMID: 31742600
- PMCID: PMC6862929
- DOI: 10.1093/gigascience/giz132
RepeatFiller newly identifies megabases of aligning repetitive sequences and improves annotations of conserved non-exonic elements
Abstract
Background: Transposons and other repetitive sequences make up a large part of complex genomes. Repetitive sequences can be co-opted into a variety of functions and thus provide a source for evolutionary novelty. However, comprehensively detecting ancestral repeats that align between species is difficult because considering all repeat-overlapping seeds in alignment methods that rely on the seed-and-extend heuristic results in prohibitively high runtimes.
Results: Here, we show that ignoring repeat-overlapping alignment seeds when aligning entire genomes misses numerous alignments between repetitive elements. We present a tool, RepeatFiller, that improves genome alignments by incorporating previously undetected local alignments between repetitive sequences. By applying RepeatFiller to genome alignments between human and 20 other representative mammals, we uncover between 22 and 84 Mb of previously undetected alignments that mostly overlap transposable elements. We further show that the increased alignment coverage improves the annotation of conserved non-exonic elements, both by discovering numerous novel transposon-derived elements that evolve under constraint and by removing thousands of elements that are not under constraint in placental mammals.
Conclusions: RepeatFiller contributes to comprehensively aligning repetitive genomic regions, which facilitates studying transposon co-option and genome evolution. Source code: https://github.com/hillerlab/GenomeAlignmentTools.
Keywords: conserved non-exonic elements; genome alignments; transposons.
© The Author(s) 2019. Published by Oxford University Press.
Figures


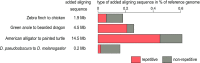



Similar articles
-
A genome alignment of 120 mammals highlights ultraconserved element variability and placenta-associated enhancers.Gigascience. 2020 Jan 1;9(1):giz159. doi: 10.1093/gigascience/giz159. Gigascience. 2020. PMID: 31899510 Free PMC article.
-
Distribution and intensity of constraint in mammalian genomic sequence.Genome Res. 2005 Jul;15(7):901-13. doi: 10.1101/gr.3577405. Epub 2005 Jun 17. Genome Res. 2005. PMID: 15965027 Free PMC article.
-
Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome.Genome Res. 2007 Jun;17(6):760-74. doi: 10.1101/gr.6034307. Genome Res. 2007. PMID: 17567995 Free PMC article.
-
Use of long sequence alignments to study the evolution and regulation of mammalian globin gene clusters.Mol Biol Evol. 1993 Jan;10(1):73-102. doi: 10.1093/oxfordjournals.molbev.a039991. Mol Biol Evol. 1993. PMID: 8383794 Review.
-
Repetitive sequences in complex genomes: structure and evolution.Annu Rev Genomics Hum Genet. 2007;8:241-59. doi: 10.1146/annurev.genom.8.080706.092416. Annu Rev Genomics Hum Genet. 2007. PMID: 17506661 Review.
Cited by
-
Contradictory Phylogenetic Signals in the Laurasiatheria Anomaly Zone.Genes (Basel). 2022 Apr 26;13(5):766. doi: 10.3390/genes13050766. Genes (Basel). 2022. PMID: 35627151 Free PMC article.
-
Interspecies transcriptomics identify genes that underlie disproportionate foot growth in jerboas.Curr Biol. 2022 Jan 24;32(2):289-303.e6. doi: 10.1016/j.cub.2021.10.063. Epub 2021 Nov 17. Curr Biol. 2022. PMID: 34793695 Free PMC article.
-
Finding and Characterizing Repeats in Plant Genomes.Methods Mol Biol. 2022;2443:327-385. doi: 10.1007/978-1-0716-2067-0_18. Methods Mol Biol. 2022. PMID: 35037215
-
Distinct Genes with Similar Functions Underlie Convergent Evolution in Myotis Bat Ecomorphs.Mol Biol Evol. 2024 Sep 4;41(9):msae165. doi: 10.1093/molbev/msae165. Mol Biol Evol. 2024. PMID: 39116340 Free PMC article.
-
Chromosome-length genome assembly and linkage map of a critically endangered Australian bird: the helmeted honeyeater.Gigascience. 2022 Mar 29;11:giac025. doi: 10.1093/gigascience/giac025. Gigascience. 2022. PMID: 35348671 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

