Statistically Driven Metabolite and Lipid Profiling of Patients from the Undiagnosed Diseases Network
- PMID: 31742994
- PMCID: PMC7183858
- DOI: 10.1021/acs.analchem.9b03522
Statistically Driven Metabolite and Lipid Profiling of Patients from the Undiagnosed Diseases Network
Abstract
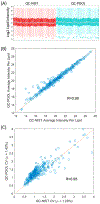
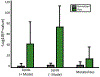
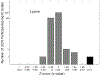
Advancements in molecular separations coupled with mass spectrometry have enabled metabolome analyses for clinical cohorts. A population of interest for metabolome profiling is patients with rare disease for which abnormal metabolic signatures may yield clues into the genetic basis, as well as mechanistic drivers of the disease and possible treatment options. We undertook the metabolome profiling of a large cohort of patients with mysterious conditions characterized through the Undiagnosed Diseases Network (UDN). Due to the size and enrollment procedures, collection of the metabolomes for UDN patients took place over 2 years. We describe the study designed to adjust for measurements collected over a long time scale and how this enabled statistical analyses to summarize the metabolome of individual patients. We demonstrate the removal of time-based batch effects, overall statistical characteristics of the UDN population, and two case studies of interest that demonstrate the utility of metabolome profiling for rare diseases.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

