Human induced pluripotent stem cells from two azoospermic patients with Klinefelter syndrome show similar X chromosome inactivation behavior to female pluripotent stem cells
- PMID: 31743397
- PMCID: PMC6894010
- DOI: 10.1093/humrep/dez134
Human induced pluripotent stem cells from two azoospermic patients with Klinefelter syndrome show similar X chromosome inactivation behavior to female pluripotent stem cells
Abstract
Study question: Does the X chromosome inactivation (XCI) of Klinefelter syndrome (KS)-derived human induced pluripotent stem cells (hiPSCs) correspond to female human pluripotent stem cells (hPSCs) and reflect the KS genotype?
Summary answer: Our results demonstrate for the first time that KS-derived hiPSCs show similar XCI behavior to female hPSCs in culture and show biological relevance to KS genotype-related clinical features.
What is known already: So far, assessment of XCI of KS-derived hiPSCs was based on H3K27me3 staining and X-inactive specific transcript gene expression disregarding the at least three XCI states (XaXi with XIST coating, XaXi lacking XIST coating, and XaXe (partially eroded XCI)) that female hPSCs display in culture.
Study design, size, duration: The study used hiPSC lines generated from two azoospermic patients with KS and included two healthy male (HM) and one healthy female donor.
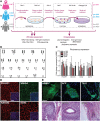
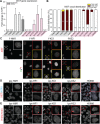
Participants/materials, setting, methods: In this study, we derived hiPSCs by reprograming fibroblasts with episomal plasmids and applying laminin 521 as culture substrate. hiPSCs were characterized by karyotyping, immunocytochemistry, immunohistochemistry, quantitative PCR, teratoma formation, and embryoid body differentiation. XCI and KS hiPSC relevance were assessed by whole genome transcriptomics analysis and immunocytochemistry plus FISH of KS, HM and female fibroblast, and their hiPSC derivatives.
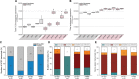
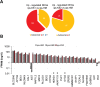
Main results and the role of chance: Applying whole genome transcriptomics analysis, we could identify differentially expressed genes (DEGs) between KS and HM donors with enrichment in gene ontology terms associated with fertility, cardiovascular development, ossification, and brain development, all associated with KS genotype-related clinical features. Furthermore, XCI analysis based on transcriptomics data, RNA FISH, and H3K27me3 staining revealed variable XCI states of KS hiPSCs similar to female hiPSCs, showing either normal (XaXi) or eroded (XaXe) XCI. KS hiPSCs with normal XCI showed nevertheless upregulated X-linked genes involved in nervous system development as well as synaptic transmission, supporting the potential use of KS-derived hiPSCs as an in vitro model for KS.
Limitations, reasons for caution: Detailed clinical information for patients included in this study was not available. Although a correlation between DEGs and the KS genotype could be observed, the biological relevance of these cells has to be confirmed with further experiments. In addition, karyotype analysis for two hiPSC lines was performed at passage 12 but not repeated at a later passage. Nevertheless, since all XCI experiments for those lines were performed between passage 11 and 15 the authors expect no karyotypic changes for those experiments.
Wider implications of the findings: As KS patients have variable clinical phenotypes that are influenced by the grade of aneuploidy, mosaicism, origin of the X chromosome, and XCI 'escapee' genes, which vary not only among individuals but also among different tissues within the same individual, differentiated KS hiPSCs could be used for a better understanding of KS pathogenesis.
Study funding/competing interest(s): This study was supported by grants from the Knut and Alice Wallenberg Foundation (2016.0121 and 2015.0096), Ming Wai Lau Centre for Reparative Medicine (2-343/2016), Ragnar Söderberg Foundation (M67/13), Swedish Research Council (2013-32485-100360-69), the Centre for Innovative Medicine (2-388/2016-40), Kronprinsessan Lovisas Förening För Barnasjukvård/Stiftelsen Axel Tielmans Minnesfond, Samariten Foundation, Jonasson Center at the Royal Institute of Technology, Sweden, and Initial Training Network Marie Curie Program 'Growsperm' (EU-FP7-PEOPLE-2013-ITN 603568). The authors declare no conflicts of interest.
Keywords: Klinefelter syndrome; X chromosome inactivation; XXY condition; human induced pluripotent stem cells; integration-free; laminin 521; xeno-free.
© The Author(s) 2019. Published by Oxford University Press on behalf of the European Society of Human Reproduction and Embryology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

