Structural and functional modularity of the U2 snRNP in pre-mRNA splicing
- PMID: 31744343
- PMCID: PMC6934263
- DOI: 10.1080/10409238.2019.1691497
Structural and functional modularity of the U2 snRNP in pre-mRNA splicing
Abstract
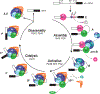
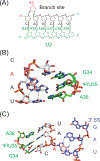
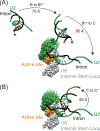
The U2 small nuclear ribonucleoprotein (snRNP) is an essential component of the spliceosome, the cellular machine responsible for removing introns from precursor mRNAs (pre-mRNAs) in all eukaryotes. U2 is an extraordinarily dynamic splicing factor and the most frequently mutated in cancers. Cryo-electron microscopy (cryo-EM) has transformed our structural and functional understanding of the role of U2 in splicing. In this review, we synthesize these and other data with respect to a view of U2 as an assembly of interconnected functional modules. These modules are organized by the U2 small nuclear RNA (snRNA) for roles in spliceosome assembly, intron substrate recognition, and protein scaffolding. We describe new discoveries regarding the structure of U2 components and how the snRNP undergoes numerous conformational and compositional changes during splicing. We specifically highlight large scale movements of U2 modules as the spliceosome creates and rearranges its active site. U2 serves as a compelling example for how cellular machines can exploit the modular organization and structural plasticity of an RNP.
Keywords: Pre-mRNA; RNA; SF3B1; snRNP; spliceosome; splicing.
Conflict of interest statement
Disclosure statement
The authors have no potential conflicts of interest to report.
Figures

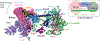

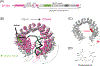
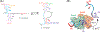


References
-
- Abovich N, Liao XLC, Rosbash M. 1994. The yeast Mud2 protein - An interaction with Prp11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 8(7):843–854. - PubMed
-
- Agrawal AA, Yu L, Smith PG, Buonamici S. 2018. Targeting splicing abnormalities in cancer. Current opinion in genetics & development. 48:67–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
