The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens
- PMID: 31744546
- PMCID: PMC6864930
- DOI: 10.1186/s13059-019-1835-8
The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens
Abstract
Background: The Critical Assessment of Functional Annotation (CAFA) is an ongoing, global, community-driven effort to evaluate and improve the computational annotation of protein function.
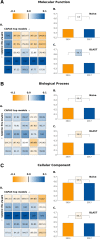
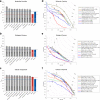
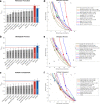
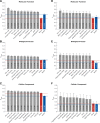
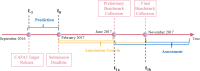
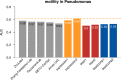
Results: Here, we report on the results of the third CAFA challenge, CAFA3, that featured an expanded analysis over the previous CAFA rounds, both in terms of volume of data analyzed and the types of analysis performed. In a novel and major new development, computational predictions and assessment goals drove some of the experimental assays, resulting in new functional annotations for more than 1000 genes. Specifically, we performed experimental whole-genome mutation screening in Candida albicans and Pseudomonas aureginosa genomes, which provided us with genome-wide experimental data for genes associated with biofilm formation and motility. We further performed targeted assays on selected genes in Drosophila melanogaster, which we suspected of being involved in long-term memory.
Conclusion: We conclude that while predictions of the molecular function and biological process annotations have slightly improved over time, those of the cellular component have not. Term-centric prediction of experimental annotations remains equally challenging; although the performance of the top methods is significantly better than the expectations set by baseline methods in C. albicans and D. melanogaster, it leaves considerable room and need for improvement. Finally, we report that the CAFA community now involves a broad range of participants with expertise in bioinformatics, biological experimentation, biocuration, and bio-ontologies, working together to improve functional annotation, computational function prediction, and our ability to manage big data in the era of large experimental screens.
Keywords: Biofilm; Community challenge; Critical assessment; Long-term memory; Protein function prediction.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

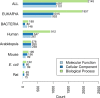
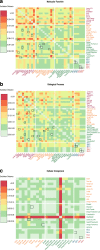


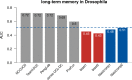
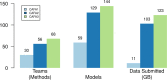


References
Publication types
MeSH terms
Grants and funding
- BB/M025047/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- P20 GM113132/GM/NIGMS NIH HHS/United States
- R00 GM097033/GM/NIGMS NIH HHS/United States
- BB/N019431/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R21 NS103831/NS/NINDS NIH HHS/United States
- U01 CA198942/CA/NCI NIH HHS/United States
- U41 HG007234/HG/NHGRI NIH HHS/United States
- BB/F00964X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- U24 TR002306/TR/NCATS NIH HHS/United States
- R35 GM128637/GM/NIGMS NIH HHS/United States
- BB/L020505/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 GM093123/GM/NIGMS NIH HHS/United States
- MC_UP_1201/14/MRC_/Medical Research Council/United Kingdom
- R01 GM071749/GM/NIGMS NIH HHS/United States
- R01 GM123055/GM/NIGMS NIH HHS/United States
- R15 GM120650/GM/NIGMS NIH HHS/United States
- BB/N004876/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L002817/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/K004131/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M015009/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- UL1 TR002319/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

