Towards scaling elementary flux mode computation
- PMID: 31745550
- PMCID: PMC8499997
- DOI: 10.1093/bib/bbz094
Towards scaling elementary flux mode computation
Abstract
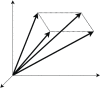
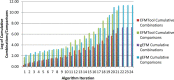
While elementary flux mode (EFM) analysis is now recognized as a cornerstone computational technique for cellular pathway analysis and engineering, EFM application to genome-scale models remains computationally prohibitive. This article provides a review of aspects of EFM computation that elucidates bottlenecks in scaling EFM computation. First, algorithms for computing EFMs are reviewed. Next, the impact of redundant constraints, sensitivity to constraint ordering and network compression are evaluated. Then, the advantages and limitations of recent parallelization and GPU-based efforts are highlighted. The article then reviews alternative pathway analysis approaches that aim to reduce the EFM solution space. Despite advances in EFM computation, our review concludes that continued scaling of EFM computation is necessary to apply EFM to genome-scale models. Further, our review concludes that pathway analysis methods that target specific pathway properties can provide powerful alternatives to EFM analysis.
Keywords: elementary flux mode analysis; elementary flux modes; parallel computing; pathway analysis; scalability.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
References
-
- Aldor IS, Keasling JD. Process design for microbial plastic factories: metabolic engineering of polyhydroxyalkanoates. Curr Opin Biotechnol 2003;14(5):475–83. - PubMed
-
- Nakamura CE, Whited GM. Metabolic engineering for the microbial production of 1,3-propanediol. Curr Opin Biotechnol 2003;14(5):454–9. - PubMed
-
- Steen EJ, Kang Y, Bokinsky G, et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 2010;463(7280):559–62. - PubMed
-
- Chang MC, Keasling JD. Production of isoprenoid pharmaceuticals by engineered microbes. Nat Chem Biol 2006;2(12):674–81. - PubMed
-
- Martin VJ, Pitera DJ, Withers ST, et al. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 2003;21(7):796–802. - PubMed

