Apoptotic SKOV3 cells stimulate M0 macrophages to differentiate into M2 macrophages and promote the proliferation and migration of ovarian cancer cells by activating the ERK signaling pathway
- PMID: 31746376
- PMCID: PMC6889918
- DOI: 10.3892/ijmm.2019.4408
Apoptotic SKOV3 cells stimulate M0 macrophages to differentiate into M2 macrophages and promote the proliferation and migration of ovarian cancer cells by activating the ERK signaling pathway
Abstract
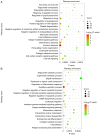
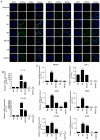
Ovarian cancer has a high rate of recurrence, with M2 macrophages having been found to be involved in its progression and metastasis. To examine the relationship between macrophages and ovarian cancer in the present study, M0 macrophages were stimulated with apoptotic SKOV3 cells and it was found that these macrophages promoted tumor proliferation and migration. Subsequently, the mRNAs and proteins expressed at high levels in these M2 macrophages were examined by RNA‑Seq and quantitative proteomics, respectively, which revealed that M0 macrophages stimulated by apoptotic SKOV3 cells also expressed M2 markers, including CD206, interleukin‑10, C‑C motif chemokine ligand 22, aminopeptidase‑N, disabled homolog 2, matrix metalloproteinase 1 and 5'‑nucleotidase. The abundance of phosphorylated Erk1/2 in these macrophages was increased. The results indicate that apoptotic SKOV3 cells stimulate M0 macrophages to differentiate into M2 macrophages by activating the ERK pathway. These results suggest possible treatments for patients with ovarian cancer who undergo chemotherapy; inhibiting M2 macrophage differentiation during chemotherapy may reduce the rate of tumor recurrence.
Keywords: apoptotic SKOV3; M2 macrophage; quantitative proteomics; RNA-Seq; ERK pathway.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

