Identification of potential hub genes related to the progression and prognosis of hepatocellular carcinoma through integrated bioinformatics analysis
- PMID: 31746405
- PMCID: PMC6908929
- DOI: 10.3892/or.2019.7400
Identification of potential hub genes related to the progression and prognosis of hepatocellular carcinoma through integrated bioinformatics analysis
Abstract
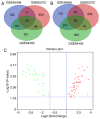
Hepatocellular carcinoma (HCC) is the fourth leading cause of cancer‑related deaths among cancer patients. Genes correlated with the progression and prognosis of HCC are critically needed to be identified. In the present study, 3 Gene Expression Omnibus (GEO) datasets (GSE46408, GSE65372 and GSE84402) were used to analyze the differentially expressed genes (DEGs) between HCC and non‑tumor liver tissues. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were conducted to clarify the functional roles of DEGs. A protein‑protein interaction network was established to screen the hub genes associated with HCC. The prognostic values of hub genes in HCC patients were analyzed using The Cancer Genome Atlas (TCGA) database. The expression levels of hub genes were validated based on ONCOMINE, TCGA and Human Protein Atlas (HPA) databases. Notably, 56 upregulated and 33 downregulated DEGs were markedly enriched under various GO terms and four KEGG terms. Among these DEGs, 10 hub genes with high connectivity degree were identified, including cyclin B1, cyclin A2, cyclin B2, condensin complex subunit 3, PDZ binding kinase, nucleolar and spindle‑associated protein 1, aurora kinase A, ZW10 interacting kinetochore protein, protein regulator of cytokinesis 1 and kinesin family member 4A. The upregulated expression levels of these hub genes in HCC tissues were further confirmed by ONCOMINE, TCGA, and HPA databases. Additionally, the increased mRNA expression of each hub gene was related to the unfavorable disease‑free survival and overall survival of HCC patients. The present study identified ten genes associated with HCC, which may help to provide candidate targets for the diagnosis and treatment of HCC.
Keywords: hepatocellular carcinoma; differently expressed genes; bioinformatics analysis; hub gene; prognosis.
Figures











References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

