Comparative genomic analysis and molecular examination of the diversity of enterotoxigenic Escherichia coli isolates from Chile
- PMID: 31747410
- PMCID: PMC6901236
- DOI: 10.1371/journal.pntd.0007828
Comparative genomic analysis and molecular examination of the diversity of enterotoxigenic Escherichia coli isolates from Chile
Abstract
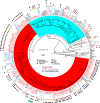
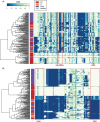
Enterotoxigenic Escherichia coli (ETEC) is one of the most common diarrheal pathogens in the low- and middle-income regions of the world, however a systematic examination of the genomic content of isolates from Chile has not yet been undertaken. Whole genome sequencing and comparative analysis of a collection of 125 ETEC isolates from three geographic locations in Chile, allowed the interrogation of phylogenomic groups, sequence types and genes specific to isolates from the different geographic locations. A total of 80.8% (101/125) of the ETEC isolates were identified in E. coli phylogroup A, 15.2% (19/125) in phylogroup B, and 4.0% (5/125) in phylogroup E. The over-representation of genomes in phylogroup A was significantly different from other global ETEC genomic studies. The Chilean ETEC isolates could be further subdivided into sub-clades similar to previously defined global ETEC reference lineages that had conserved multi-locus sequence types and toxin profiles. Comparison of the gene content of the Chilean ETEC identified genes that were unique based on geographic location within Chile, phylogenomic classifications or sequence type. Completion of a limited number of genomes provided insight into the ETEC plasmid content, which is conserved in some phylogenomic groups and not conserved in others. These findings suggest that the Chilean ETEC isolates contain unique virulence factor combinations and genomic content compared to global reference ETEC isolates.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- WHO. Future directions for research on enterotoxigenic Escherichia coli vaccines for developing countries. Weekly Epidemiological Record. 2006;81(11):97–104. - PubMed
-
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382(9888):209–22. Epub 2013/05/18. S0140-6736(13)60844-2 [pii] 10.1016/S0140-6736(13)60844-2 . - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

