The Bromodomain Protein 4 Contributes to the Regulation of Alternative Splicing
- PMID: 31747612
- PMCID: PMC6893865
- DOI: 10.1016/j.celrep.2019.10.066
The Bromodomain Protein 4 Contributes to the Regulation of Alternative Splicing
Abstract
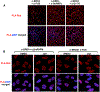
The bromodomain protein 4 (BRD4) is an atypical kinase and histone acetyl transferase (HAT) that binds to acetylated histones and contributes to chromatin remodeling and early transcriptional elongation. During transcription, BRD4 travels with the elongation complex. Since most alternative splicing events take place co-transcriptionally, we asked if BRD4 plays a role in regulating alternative splicing. We report that distinct patterns of alternative splicing are associated with a conditional deletion of BRD4 during thymocyte differentiation in vivo. Similarly, the depletion of BRD4 in T cell acute lymphoblastic leukemia (T-ALL) cells alters patterns of splicing. Most alternatively spliced events affected by BRD4 are exon skipping. Importantly, BRD4 interacts with components of the splicing machinery, as assessed by both immunoprecipitation (IP) and proximity ligation assays (PLAs), and co-localizes on chromatin with the splicing regulator, FUS. We propose that BRD4 contributes to patterns of alternative splicing through its interaction with the splicing machinery during transcription elongation.
Keywords: AML; BET; BRD4; FUS; alternative splicing; thymocyte differentiation.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures

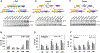



References
-
- Berkovits BD, Wang L, Guarnieri P, and Wolgemuth DJ (2012). The testis-specific double bromodomain-containing protein BRDT forms a complex with multiple spliceosome components and is required for mRNA splicing and 3′-UTR truncation in round spermatids. Nucleic Acids Res. 40, 7162–7175. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

