Novel Sulfolobus Fuselloviruses with Extensive Genomic Variations
- PMID: 31748395
- PMCID: PMC6997748
- DOI: 10.1128/JVI.01624-19
Novel Sulfolobus Fuselloviruses with Extensive Genomic Variations
Erratum in
-
Erratum for Zhang et al., "Novel Sulfolobus Fuselloviruses with Extensive Genomic Variations".J Virol. 2020 Mar 31;94(8):e00192-20. doi: 10.1128/JVI.00192-20. Print 2020 Mar 31. J Virol. 2020. PMID: 32234800 Free PMC article. No abstract available.
Abstract
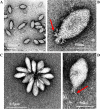
Fuselloviruses are among the most widespread and best-characterized archaeal viruses. They exhibit remarkable diversity, as the list of members of this family is rapidly growing. However, it has yet to be shown how a fuselloviral genome may undergo variation at the levels of both single nucleotides and sequence stretches. Here, we report the isolation and characterization of four novel spindle-shaped viruses, named Sulfolobus spindle-shaped viruses 19 to 22 (SSV19-22), from a hot spring in the Philippines. SSV19 is a member of the genus Alphafusellovirus, whereas SSV20-22 belong to the genus Betafusellovirus The genomes of SSV20-SSV22 are identical except for the presence of two large variable regions, as well as numerous sites of single-nucleotide polymorphisms (SNPs) unevenly distributed throughout the genomes and enriched in certain regions, including the gene encoding the putative end filament protein VP4. We show that coinfection of the host with SSV20 and SSV22 led to the formation of an SSV21-like virus, presumably through homologous recombination. In addition, large numbers of SNPs were identified in DNA sequences retrieved by PCR amplification targeting the SSV20-22 vp4 gene from the original enrichment culture, indicating the enormous diversity of SSV20-22-like viruses in the environment. The high variability of VP4 is consistent with its potential role in host recognition and binding by the virus.IMPORTANCE How a virus survives in the arms race with its host is an intriguing question. In this study, we isolated and characterized four novel fuselloviruses, named Sulfolobus spindle-shaped viruses 19 to 22 (SSV19-22). Interestingly, SSV20-22 differ primarily in two genomic regions and are apparently convertible through homologous recombination during coinfection. Moreover, sites of single-nucleotide polymorphism (SNP) were identified throughout the genomes of SSV20-22 and, notably, enriched in certain regions, including the gene encoding the putative end filament protein VP4, which is believed to be involved in host recognition and binding by the virus.
Keywords: fuselloviruses; genomic diversity; homologous recombination; single-nucleotide polymorphism; virus-host interaction.
Copyright © 2020 American Society for Microbiology.
Figures







Similar articles
-
Four newly isolated fuselloviruses from extreme geothermal environments reveal unusual morphologies and a possible interviral recombination mechanism.Environ Microbiol. 2009 Nov;11(11):2849-62. doi: 10.1111/j.1462-2920.2009.02009.x. Epub 2009 Jul 23. Environ Microbiol. 2009. PMID: 19638177
-
Extreme Mutation Tolerance: Nearly Half of the Archaeal Fusellovirus Sulfolobus Spindle-Shaped Virus 1 Genes Are Not Required for Virus Function, Including the Minor Capsid Protein Gene vp3.J Virol. 2017 Apr 28;91(10):e02406-16. doi: 10.1128/JVI.02406-16. Print 2017 May 15. J Virol. 2017. PMID: 28148789 Free PMC article.
-
Sulfolobus tengchongensis spindle-shaped virus STSV1: virus-host interactions and genomic features.J Virol. 2005 Jul;79(14):8677-86. doi: 10.1128/JVI.79.14.8677-8686.2005. J Virol. 2005. PMID: 15994761 Free PMC article.
-
Plasmids and viruses of the thermoacidophilic crenarchaeote Sulfolobus.Extremophiles. 2006 Feb;10(1):17-28. doi: 10.1007/s00792-005-0492-x. Epub 2006 Jan 6. Extremophiles. 2006. PMID: 16397749 Review.
-
Molecular biology of fuselloviruses and their satellites.Extremophiles. 2014 May;18(3):473-89. doi: 10.1007/s00792-014-0634-0. Epub 2014 Feb 23. Extremophiles. 2014. PMID: 24562787 Review.
Cited by
-
Sulfolobus islandicus Employs Orc1-2-Mediated DNA Damage Response in Defense against Infection by SSV2.J Virol. 2022 Dec 21;96(24):e0143822. doi: 10.1128/jvi.01438-22. Epub 2022 Nov 30. J Virol. 2022. PMID: 36448807 Free PMC article.
-
Isolation and Characterization of the First Temperate Virus Infecting Psychrobacillus from Marine Sediments.Viruses. 2022 Jan 8;14(1):108. doi: 10.3390/v14010108. Viruses. 2022. PMID: 35062312 Free PMC article.
-
Host-Dependent Differences in Replication Strategy of the Sulfolobus Spindle-Shaped Virus Strain SSV9 (a.k.a., SSVK1): Infection Profiles in Hosts of the Family Sulfolobaceae.Front Microbiol. 2020 Jul 14;11:1218. doi: 10.3389/fmicb.2020.01218. eCollection 2020. Front Microbiol. 2020. PMID: 32760354 Free PMC article.
-
Life in hot acid: a genome-based reassessment of the archaeal order Sulfolobales.Environ Microbiol. 2021 Jul;23(7):3568-3584. doi: 10.1111/1462-2920.15189. Epub 2020 Sep 7. Environ Microbiol. 2021. PMID: 32776389 Free PMC article.
-
An archaeal virus capable of hydrolyzing the surface glycan of the host cell.mLife. 2025 Apr 3;4(2):219-222. doi: 10.1002/mlf2.70008. eCollection 2025 Apr. mLife. 2025. PMID: 40313977 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

