Hepatitis Delta Virus Alters the Autophagy Process To Promote Its Genome Replication
- PMID: 31748400
- PMCID: PMC6997767
- DOI: 10.1128/JVI.01936-19
Hepatitis Delta Virus Alters the Autophagy Process To Promote Its Genome Replication
Abstract
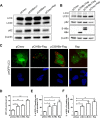
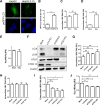
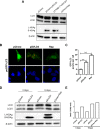
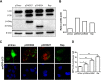
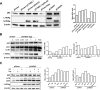
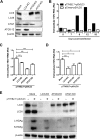
A substantial number of viruses have been demonstrated to subvert autophagy to promote their own replication. Recent publications have reported the proviral effect of autophagy induction on hepatitis B virus (HBV) replication. Hepatitis delta virus (HDV) is a defective virus and an occasional obligate satellite of HBV. However, no previous work has studied the relationship between autophagy and HDV. In this article, we analyze the impact of HBV and HDV replication on autophagy as well as the involvement of the autophagy machinery in the HDV life cycle when produced alone and in combination with HBV. We prove that HBxAg and HBsAg can induce early steps of autophagy but ultimately block flux. It is worth noting that the two isoforms of the HDV protein, the small HDAg (S-HDAg) and large HDAg (L-HDAg) isoforms, can also efficiently promote autophagosome accumulation and disturb autophagic flux. Using CRISPR-Cas9 technology to generate specific knockouts, we demonstrate that the autophagy machinery, specifically the proteins implicated in the elongation step (ATG7, ATG5, and LC3), is important for the release of HBV without affecting the level of intracellular HBV genomes. Surprisingly, the knockout of ATG5 and ATG7 decreased the intracellular HDV RNA level in both Huh7 and HepG2.2.15 cells without an additional effect on HDV secretion. Therefore, we conclude that HBV and HDV have evolved to utilize the autophagy machinery so as to assist at different steps of their life cycle.IMPORTANCE Hepatitis delta virus is a defective RNA virus that requires hepatitis B virus envelope proteins (HBsAg) to fulfill its life cycle. Thus, HDV can only infect individuals at the same time as HBV (coinfection) or superinfect individuals who are already chronic carriers of HBV. The presence of HDV in the liver accelerates the progression of infection to fibrosis and to hepatic cancer. Since current treatments against HBV are ineffective against HDV, it is of paramount importance to study the interaction between HBV, HDV, and host factors. This will help unravel new targets whereby a therapy that is capable of simultaneously impeding both viruses could be developed. In this research paper, we evidence that the autophagy machinery promotes the replication of HBV and HDV at different steps of their life cycle. Notwithstanding their contribution to HBV release, autophagy proteins seem to assist HDV intracellular replication but not its secretion.
Keywords: ATG5; autophagy; chronic infection; hepatitis B virus (HBV); hepatitis delta virus (HDV); viral replication.
Copyright © 2020 American Society for Microbiology.
Figures

Similar articles
-
An update on HDV: virology, pathogenesis and treatment.Antivir Ther. 2013;18(3 Pt B):541-8. doi: 10.3851/IMP2598. Epub 2013 Jun 21. Antivir Ther. 2013. PMID: 23792471 Review.
-
Analysis of Replication, Cell Division-Mediated Spread, and HBV Envelope Protein-Dependent Pseudotyping of Three Mammalian Delta-like Agents.Viruses. 2024 May 28;16(6):859. doi: 10.3390/v16060859. Viruses. 2024. PMID: 38932152 Free PMC article.
-
Hepatitis D virus infection, replication and cross-talk with the hepatitis B virus.World J Gastroenterol. 2014 Oct 28;20(40):14589-97. doi: 10.3748/wjg.v20.i40.14589. World J Gastroenterol. 2014. PMID: 25356023 Free PMC article.
-
A new HDV mouse model identifies mitochondrial antiviral signaling protein (MAVS) as a key player in IFN-β induction.J Hepatol. 2017 Oct;67(4):669-679. doi: 10.1016/j.jhep.2017.05.010. Epub 2017 May 18. J Hepatol. 2017. PMID: 28527664
-
Future treatments for hepatitis delta virus infection.Liver Int. 2020 Feb;40 Suppl 1:54-60. doi: 10.1111/liv.14356. Liver Int. 2020. PMID: 32077603 Review.
Cited by
-
Prevalence and molecular characterization of hepatitis delta virus infection among hepatitis B virus surface antigen positive students and pregnant women in N'djamena, Chad.IJID Reg. 2024 Dec 28;14:100560. doi: 10.1016/j.ijregi.2024.100560. eCollection 2025 Mar. IJID Reg. 2024. PMID: 39895833 Free PMC article.
-
Autophagy machinery as exploited by viruses.Autophagy Rep. 2025 Mar 18;4(1):2464986. doi: 10.1080/27694127.2025.2464986. eCollection 2025 Dec 31. Autophagy Rep. 2025. PMID: 40201908 Free PMC article.
-
Fangchinoline inhibits the PEDV replication in intestinal epithelial cells via autophagic flux suppression.Front Microbiol. 2023 Jul 7;14:1164851. doi: 10.3389/fmicb.2023.1164851. eCollection 2023. Front Microbiol. 2023. PMID: 37485535 Free PMC article.
-
Viral hepatitis update: Progress and perspectives.World J Gastroenterol. 2021 Jul 14;27(26):4018-4044. doi: 10.3748/wjg.v27.i26.4018. World J Gastroenterol. 2021. PMID: 34326611 Free PMC article. Review.
-
Exosomes as Conduits: Facilitating Hepatitis B Virus-Independent Hepatitis D Virus Transmission and Propagation in Hepatocytes.Viruses. 2024 May 22;16(6):825. doi: 10.3390/v16060825. Viruses. 2024. PMID: 38932118 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

