Genomic Signatures for Sedimentary Microbial Utilization of Phytoplankton Detritus in a Fast-Flowing Estuary
- PMID: 31749780
- PMCID: PMC6848030
- DOI: 10.3389/fmicb.2019.02475
Genomic Signatures for Sedimentary Microbial Utilization of Phytoplankton Detritus in a Fast-Flowing Estuary
Abstract
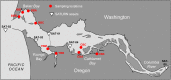
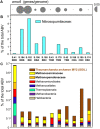
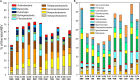
In fast-flowing, river-dominated estuaries, "hotspots" of microbial biogeochemical cycling can be found within areas of extended water retention. Lateral bays located off of the North and South channels of the Columbia River estuary are proposed to be such hotspots. Previous metagenomic studies on water samples indicated that these regions function both as sources and sinks of biogenic particles, with potential to impact organic matter fluxes in the estuary. To extend this work, we analyzed 11 sediment metagenomes from three disparate bays: the freshwater Cathlamet Bay, and the brackish Youngs Bay and more saline Baker Bay located nearer the mouth to the south and north of the main channel, respectively. Samples were collected from upper layers of sediments in August of 2011 and 2013 for DNA extraction and metagenome sequencing. All metagenomes were dominated by bacterial sequences, although diatom sequences as high as 26% of the total annotated sequences were observed in the higher salinity samples. Unsupervised 2D hierarchical clustering analysis resulted in the eleven metagenome samples clustered into four groups by microbial taxonomic composition, with Bacteroides, diatom, and phage levels driving most of the grouping. Results of functional gene clustering further indicated that diatom bloom degradation stage (early vs. late) was an important factor. While the Flavobacteriia and Cytophagia classes were well represented in metagenomes containing abundant diatoms, taxa from the Bacteroidia class, along with certain members of the Sphingobacteriia class, were particularly abundant in metagenomes representing later stages of diatom decomposition. In contrast, the sediment metagenomes with low relative abundance of diatom and Bacteroidetes sequences appeared to have a metabolic potential biased toward microbial growth under nutrient limitation. While differences in water salinity clearly also influenced the microbial community composition and metabolic potential, our results highlight a central role for allochthonous labile organic matter (i.e., diatom detritus), in shaping bacterial taxonomic and functional properties in the Columbia River estuary lateral bay sediments. These results suggest that in fast-flowing, river-dominated estuaries, sediment microbial communities in areas of extended water retention, such as the lateral bays, may contribute disproportionately to estuarine organic matter degradation and recycling.
Keywords: Bacteroidetes; bacteriophage; estuarine sediments; metagenomics; microbial community; phytoplankton.
Copyright © 2019 Smith, Herfort, Rivers and Simon.
Figures







References
-
- Banuelos M. A., Klein R. D., Alexander-Bowman S. J., Rodriguez-Navarro A. (1995). A potassium transporter of the yeast Schwanniomyces occidentalis homologous to the Kup system of Escherichia coli has a high concentrative capacity. EMBO J. 14 3021–3027. 10.1002/j.1460-2075.1995.tb07304.x - DOI - PMC - PubMed
-
- Baptista A. M., Seaton C., Wilkin M. P., Riseman S. F., Needoba J. A., Maier D., et al. (2015). Infrastructure for collaborative science and societal applications in the Columbia River estuary. Front. Earth Sci. 9 659–682. 10.1007/s11707-015-0540-5 - DOI
LinkOut - more resources
Full Text Sources

