The NF-κB RelA Transcription Factor Is Critical for Regulatory T Cell Activation and Stability
- PMID: 31749798
- PMCID: PMC6842949
- DOI: 10.3389/fimmu.2019.02487
The NF-κB RelA Transcription Factor Is Critical for Regulatory T Cell Activation and Stability
Abstract
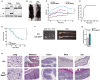
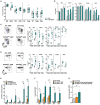
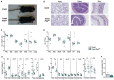
Regulatory T cells (Tregs) play a major role in immune homeostasis and in the prevention of autoimmune diseases. It has been shown that c-Rel is critical in Treg thymic differentiation, but little is known on the role of NF-κB on mature Treg biology. We thus generated mice with a specific knockout of RelA, a key member of NF-κB, in Tregs. These mice developed a severe autoimmune syndrome with multi-organ immune infiltration and high activation of lymphoid and myeloid cells. Phenotypic and transcriptomic analyses showed that RelA is critical in the acquisition of the effector Treg state independently of surrounding inflammatory environment. Unexpectedly, RelA-deficient Tregs also displayed reduced stability and cells that had lost Foxp3 produced inflammatory cytokines. Overall, we show that RelA is critical for Treg biology as it promotes both the generation of their effector phenotype and the maintenance of their identity.
Keywords: NF-κB; activation; autoimmunity; regulatory T cells; relA; stability.
Copyright © 2019 Ronin, Lubrano di Ricco, Vallion, Divoux, Kwon, Grégoire, Collares, Rouers, Baud, Benoist and Salomon.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

