Maturation of Gut Microbiota and Circulating Regulatory T Cells and Development of IgE Sensitization in Early Life
- PMID: 31749800
- PMCID: PMC6842923
- DOI: 10.3389/fimmu.2019.02494
Maturation of Gut Microbiota and Circulating Regulatory T Cells and Development of IgE Sensitization in Early Life
Abstract
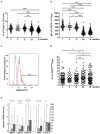
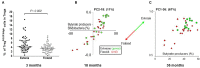
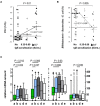
Recent studies suggest that the cross-talk between the gut microbiota and human immune system during the first year of life is an important regulator of the later development of atopic diseases. We explored the changes in the gut microbiota, blood regulatory T cells, and atopic sensitization in a birth-cohort of Estonian and Finnish children followed from 3 to 36 months of age. We describe here an infant Treg phenotype characterized by high Treg frequency, the maturation of Treg population characterized by a decrease in their frequency accompanied with an increase in the highly activated Treg cells. These changes in Treg population associated first with the relative abundance of Bifidobacterium longum followed by increasing colonization with butyrate producing bacteria. High bifidobacterial abundance in the neonatal microbiota appeared to be protective, while colonization with Bacteroides and E. coli was associated with later risk of allergy. Estonian children with lower risk of IgE mediated allergic diseases than Finnish children showed an earlier maturation of the gut microbiota, detected as earlier switch to an increasing abundance of butyrate-producing bacteria, combined with an earlier maturation of Treg cell phenotype and total IgE production. The children with established allergic diseases by age 3 showed a decreased abundance of butyrate producing Faecalibacterium. These results suggest that as well as the maintenance of a bifidobacterial dominated gut microbiota is important during the first weeks of life, the overtake by butyrate producing bacteria seems to be a beneficial shift, which should not be postponed.
Keywords: IgE; atopic diseases; bifidobacteria; gut microbiome; regulatory T-cells.
Copyright © 2019 Ruohtula, de Goffau, Nieminen, Honkanen, Siljander, Hämäläinen, Peet, Tillmann, Ilonen, Niemelä, Welling, Knip, Harmsen and Vaarala.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

