Genomic Regions Associated With Gestation Length Detected Using Whole-Genome Sequence Data Differ Between Dairy and Beef Cattle
- PMID: 31749838
- PMCID: PMC6848454
- DOI: 10.3389/fgene.2019.01068
Genomic Regions Associated With Gestation Length Detected Using Whole-Genome Sequence Data Differ Between Dairy and Beef Cattle
Abstract
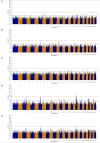
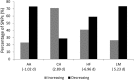
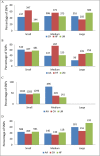
While many association studies exist that have attempted to relate genomic markers to phenotypic performance in cattle, very few have considered gestation length as a phenotype, and of those that did, none used whole genome sequence data from multiple breeds. The objective of the present study was therefore to relate imputed whole genome sequence data to estimated breeding values for gestation length using 22,566 sires (representing 2,262,706 progeny) of multiple breeds [Angus (AA), Charolais (CH), Holstein-Friesian (HF), and Limousin (LM)]. The associations were undertaken within breed using linear mixed models that accounted for genomic relatedness among sires; a separate association analysis was undertaken with all breeds analysed together but with breed included as a fixed effect in the model. Furthermore, the genome was divided into 500 kb segments and whether or not segments harboured a single nucleotide polymorphism (SNP) with a P ≤ 1 × 10-4 common to different combinations of breeds was determined. Putative quantitative trait loci (QTL) regions associated with gestation length were detected in all breeds; significant associations with gestation length were only detected in the HF population and in the across-breed analysis of all 22,566 sires. Twenty-five SNPs were significantly associated (P ≤ 5 × 10-8) with gestation length in the HF population. Of the 25 significant SNPs, 18 were located within three QTLs on Bos taurus autosome number (BTA) 18, six were in two QTL on BTA19, and one was located within a QTL on BTA7. The strongest association was rs381577268, a downstream variant of ZNF613 located within a QTL spanning from 58.06 to 58.19 Mb on BTA18; it accounted for 1.37% of the genetic variance in gestation length. Overall there were 11 HF animals within the edited dataset that were homozygous for the T allele at rs381577268 and these had a 3.3 day longer (P < 0.0001) estimated breeding value (EBV) for gestation length than the heterozygous animals and a 4.7 day longer (P < 0.0001) EBV for gestation length than the homozygous CC animals. The majority of the 500 kb windows harboring a SNP with a P ≤ 1 × 10-4 were unique to a single breed and no window was shared among all four breeds for gestation length, suggesting any QTLs identified are breed-specific associations.
Keywords: bioinformatics; genome-wide association; gestation; mixed model; pregnancy; sequence; single nucleotide polymorphism.
Copyright © 2019 Purfield, Evans, Carthy and Berry.
Figures

References
-
- Babar M. E. (2008). Heritability estimate of ewe traits in Lohi sheep, Acta Agriculturae Scandinavica. Sec. A — Anim. Sci. 58 (2), 61–64. 10.1080/09064700802137108 - DOI
-
- Berry D. P., Evans R. D. (2014). Genetics of reproductive performance in seasonal calving beef cows and its association with performance traits. J. Anim. Sci. 92 (4), 1412–1422. 10.2527 - PubMed
-
- Berry D. P., Kearney J. F., Twomey K., Evans R. D. (2013). Genetics of reproductive performance in seasonal calving dairy cattle production systems. Irish J. Agric. Food Res. 52, 1–16.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

