SMARCA4-Deficient Thoracic Sarcomatoid Tumors Represent Primarily Smoking-Related Undifferentiated Carcinomas Rather Than Primary Thoracic Sarcomas
- PMID: 31751681
- PMCID: PMC7556987
- DOI: 10.1016/j.jtho.2019.10.023
SMARCA4-Deficient Thoracic Sarcomatoid Tumors Represent Primarily Smoking-Related Undifferentiated Carcinomas Rather Than Primary Thoracic Sarcomas
Abstract
Introduction: Highly aggressive thoracic neoplasms characterized by SMARCA4 (BRG1) deficiency and undifferentiated round cell or rhabdoid morphology have been recently described and proposed to represent thoracic sarcomas. However, it remains unclear whether such tumors may instead represent sarcomatoid carcinomas, and how their clinicopathologic characteristics compare with those of nonsarcomatoid SMARCA4-deficient non-small cell lung carcinomas (SD-NSCC).
Methods: We identified 22 SMARCA4-deficient thoracic sarcomatoid tumors (SD-TSTs) with round cell and/or rhabdoid morphology and 45 SD-NSCCs, and comprehensively analyzed their clinicopathologic, immunohistochemical, and genomic characteristics using 341-468 gene next-generation sequencing and other molecular platforms.
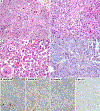
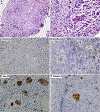
Results: The relationship of SD-TSTs with NSCC was supported by (1) the presence of NSCC components juxtaposed with sarcomatoid areas in five cases, (2) focal expression of NSCC lineage markers TTF1 or p40 in four additional cases, (3) smoking history in all except one patient (mean = 51 pack-years), accompanied by genomic smoking signature, and (4) high tumor mutation burden (mean = 14.2 mutations per megabase) and mutations characteristic of NSCC in a subset. Compared with SD-NSCCs, SD-TSTs exhibited considerably larger primary tumor size (p < 0.0001), worse survival (p = 0.004), and more frequent presentation at younger age (30-50 years) despite heavier smoking history. Distinctive pathologic features of SD-TSTs included consistent lack of adhesion molecule claudin-4, SMARCA2 (BRM) codeficiency, and frequent expression of stem cell markers.
Conclusions: SD-TSTs represent primarily smoking-associated undifferentiated/de-differentiated carcinomas rather than primary thoracic sarcomas. Despite their histogenetic relationship with NSCC, these tumors have unique clinicopathologic characteristics, supporting their recognition as a distinct entity. Further studies are warranted to determine therapeutic approaches to this novel class of exceptionally aggressive thoracic tumors.
Keywords: BRG1; Lung; Rhabdoid; SMARCA4; Sarcomatoid; Thoracic.
Copyright © 2019 International Association for the Study of Lung Cancer. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosure: The authors have no relevant conflicts of interest pertaining to this article.
Figures
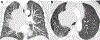



References
-
- Roberts CW, Orkin SH. The SWI/SNF complex–chromatin and cancer. Nat Rev Cancer. 2004;4:133–142. - PubMed
-
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. - PubMed
-
- Arnaud O, Le Loarer F, Tirode F. BAFfling pathologies: alterations of BAF complexes in cancer. Cancer Lett. 2018;419:266–279. - PubMed
-
- Hollmann TJ, Hornick JL. INI1-deficient tumors: diagnostic features and molecular genetics. Am J Surg Pathol. 2011;35:e47–e63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

