Recent insights into the structure of TFIID, its assembly, and its binding to core promoter
- PMID: 31751889
- PMCID: PMC7156316
- DOI: 10.1016/j.sbi.2019.10.001
Recent insights into the structure of TFIID, its assembly, and its binding to core promoter
Abstract
TFIID is a large multiprotein assembly that serves as a general transcription factor for transcription initiation by eukaryotic RNA polymerase II (Pol II). TFIID is involved in the recognition of the core promoter sequences and neighboring chromatin marks, and can interact with gene-specific activators and repressors. In order to obtain a better molecular and mechanistic understanding of the function of TFIID, its structure has been pursued for many years. However, the scarcity of TFIID and its highly flexible nature have made this pursuit very challenging. Recent breakthroughs, largely due to methodological advances in cryo-electron microscopy, have finally described the structure of this complex, both alone and engaged with core promoter DNA, revealing the functional significance of its conformational complexity in the process of core promoter recognition and initiation of Pol II transcription. Here, we review these recent structural insights and discuss their implications for our understanding of eukaryotic transcription initiation.
Copyright © 2019 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures




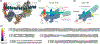
References
-
- Roeder RG: The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci 1996, 21:327–335. - PubMed
-
- Goodrich JA, Cutler G, Tjian R: Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell 1996, 84:825–830. - PubMed
-
- Burley SK, Roeder RG: Biochemistry and structural biology of transcription factor IID (TFIID). Annu Rev Biochem 1996, 65:769–799. - PubMed
-
- Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, Papadopoulos P, Tora L, Schultz P, Berger I: The architecture of human general transcription factor TFIID core complex. Nature 2013, 493:699–702. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

