A qualitative transcriptional signature for the histological reclassification of lung squamous cell carcinomas and adenocarcinomas
- PMID: 31752667
- PMCID: PMC6868745
- DOI: 10.1186/s12864-019-6086-2
A qualitative transcriptional signature for the histological reclassification of lung squamous cell carcinomas and adenocarcinomas
Abstract
Background: Targeted therapy for non-small cell lung cancer is histology dependent. However, histological classification by routine pathological assessment with hematoxylin-eosin staining and immunostaining for poorly differentiated tumors, particularly those from small biopsies, is still challenging. Additionally, the effectiveness of immunomarkers is limited by technical inconsistencies of immunostaining and lack of standardization for staining interpretation.
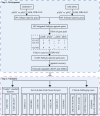
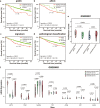
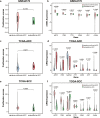
Results: Using gene expression profiles of pathologically-determined lung adenocarcinomas and squamous cell carcinomas, denoted as pADC and pSCC respectively, we developed a qualitative transcriptional signature, based on the within-sample relative gene expression orderings (REOs) of gene pairs, to distinguish ADC from SCC. The signature consists of two genes, KRT5 and AGR2, which has the stable REO pattern of KRT5 > AGR2 in pSCC and KRT5 < AGR2 in pADC. In the two test datasets with relative unambiguous NSCLC types, the apparent accuracy of the signature were 94.44 and 98.41%, respectively. In the other integrated dataset for frozen tissues, the signature reclassified 4.22% of the 805 pADC patients as SCC and 12% of the 125 pSCC patients as ADC. Similar results were observed in the clinical challenging cases, including FFPE specimens, mixed tumors, small biopsy specimens and poorly differentiated specimens. The survival analyses showed that the pADC patients reclassified as SCC had significantly shorter overall survival than the signature-confirmed pADC patients (log-rank p = 0.0123, HR = 1.89), consisting with the knowledge that SCC patients suffer poor prognoses than ADC patients. The proliferative activity, subtype-specific marker genes and consensus clustering analyses also supported the correctness of our signature.
Conclusions: The non-subjective qualitative REOs signature could effectively distinguish ADC from SCC, which would be an auxiliary test for the pathological assessment of the ambiguous cases.
Keywords: Histological subtype; Non-small cell lung cancer; Pathological assessment; Qualitative transcriptional signature; Relative gene expression orderings.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Girard L, Rodriguez-Canales J, Behrens C, Thompson DM, Botros IW, Tang H, Xie Y, Rekhtman N, Travis WD, Wistuba II, et al. An expression signature as an aid to the histologic classification of non-small cell lung cancer. Clin Cancer Res. 2016;22(19):4880–4889. doi: 10.1158/1078-0432.CCR-15-2900. - DOI - PMC - PubMed
-
- Montezuma D, Azevedo R, Lopes P, Vieira R, Cunha AL, Henrique R. A panel of four immunohistochemical markers (CK7, CK20, TTF-1, and p63) allows accurate diagnosis of primary and metastatic lung carcinoma on biopsy specimens. Virchows Arch. 2013;463(6):749–754. doi: 10.1007/s00428-013-1488-z. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- 61701143/National Natural Science Foundation of China
- 81372213/National Natural Science Foundation of China
- 81572935/National Natural Science Foundation of China
- 2016Y9044/the Joint Scientific and Technology Innovation Fund of Fujian Province
- 2017JCZX48/the Fundamental Research Funds for the Provincial Universities
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

