Multiple transcription factors co-regulate the Mycobacterium tuberculosis adaptation response to vitamin C
- PMID: 31752669
- PMCID: PMC6868718
- DOI: 10.1186/s12864-019-6190-3
Multiple transcription factors co-regulate the Mycobacterium tuberculosis adaptation response to vitamin C
Abstract
Background: Latent tuberculosis infection is attributed in part to the existence of Mycobacterium tuberculosis in a persistent non-replicating dormant state that is associated with tolerance to host defence mechanisms and antibiotics. We have recently reported that vitamin C treatment of M. tuberculosis triggers the rapid development of bacterial dormancy. Temporal genome-wide transcriptome analysis has revealed that vitamin C-induced dormancy is associated with a large-scale modulation of gene expression in M. tuberculosis.
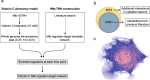
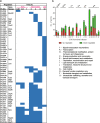
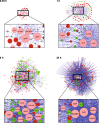
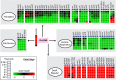
Results: An updated transcriptional regulatory network of M.tuberculosis (Mtb-TRN) consisting of 178 regulators and 3432 target genes was constructed. The temporal transcriptome data generated in response to vitamin C was overlaid on the Mtb-TRN (vitamin C Mtb-TRN) to derive insights into the transcriptional regulatory features in vitamin C-adapted bacteria. Statistical analysis using Fisher's exact test predicted that 56 regulators play a central role in modulating genes which are involved in growth, respiration, metabolism and repair functions. Rv0348, DevR, MprA and RegX3 participate in a core temporal regulatory response during 0.25 h to 8 h of vitamin C treatment. Temporal network analysis further revealed Rv0348 to be the most prominent hub regulator with maximum interactions in the vitamin C Mtb-TRN. Experimental analysis revealed that Rv0348 and DevR proteins interact with each other, and this interaction results in an enhanced binding of DevR to its target promoter. These findings, together with the enhanced expression of devR and Rv0348 transcriptional regulators, indicate a second-level regulation of target genes through transcription factor- transcription factor interactions.
Conclusions: Temporal regulatory analysis of the vitamin C Mtb-TRN revealed that there is involvement of multiple regulators during bacterial adaptation to dormancy. Our findings suggest that Rv0348 is a prominent hub regulator in the vitamin C model and large-scale modulation of gene expression is achieved through interactions of Rv0348 with other transcriptional regulators.
Keywords: Dormancy; Mtb-TRN (Mtb-transcriptional regulatory network); Mycobacterium tuberculosis; Transcriptome; Vitamin C.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Mycobacterium tuberculosis transcriptional adaptation, growth arrest and dormancy phenotype development is triggered by vitamin C.PLoS One. 2010 May 27;5(5):e10860. doi: 10.1371/journal.pone.0010860. PLoS One. 2010. PMID: 20523728 Free PMC article.
-
The pleiotropic transcriptional response of Mycobacterium tuberculosis to vitamin C is robust and overlaps with the bacterial response to multiple intracellular stresses.Microbiology (Reading). 2015 Apr;161(Pt 4):739-53. doi: 10.1099/mic.0.000049. Epub 2015 Feb 2. Microbiology (Reading). 2015. PMID: 25645949
-
Sustained expression of DevR/DosR during long-term hypoxic culture of Mycobacterium tuberculosis.Tuberculosis (Edinb). 2017 Sep;106:33-37. doi: 10.1016/j.tube.2017.06.003. Epub 2017 Jun 27. Tuberculosis (Edinb). 2017. PMID: 28802402
-
Modeling of Mycobacterium tuberculosis dormancy in bacterial cultures.Tuberculosis (Edinb). 2019 Jul;117:7-17. doi: 10.1016/j.tube.2019.05.005. Epub 2019 May 23. Tuberculosis (Edinb). 2019. PMID: 31378272 Review.
-
Adaptive Mechanisms of Mycobacterium tuberculosis: Role of fbiC Mutations in Dormancy and Survival.Int J Mycobacteriol. 2024 Oct 1;13(4):355-361. doi: 10.4103/ijmy.ijmy_198_24. Epub 2024 Dec 19. Int J Mycobacteriol. 2024. PMID: 39700156 Review.
Cited by
-
DNA structural properties of DNA binding sites for 21 transcription factors in the mycobacterial genome.Front Cell Infect Microbiol. 2023 Jun 16;13:1147544. doi: 10.3389/fcimb.2023.1147544. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37396305 Free PMC article.
-
Understanding the Reciprocal Interplay Between Antibiotics and Host Immune System: How Can We Improve the Anti-Mycobacterial Activity of Current Drugs to Better Control Tuberculosis?Front Immunol. 2021 Jun 28;12:703060. doi: 10.3389/fimmu.2021.703060. eCollection 2021. Front Immunol. 2021. PMID: 34262571 Free PMC article. Review.
-
SigE: A master regulator of Mycobacterium tuberculosis.Front Microbiol. 2023 Mar 7;14:1075143. doi: 10.3389/fmicb.2023.1075143. eCollection 2023. Front Microbiol. 2023. PMID: 36960291 Free PMC article. Review.
-
The dawn of repurposing vitamins as potential novel antimicrobial agents: A call for global emergency response amidst AMR crisis.Health Sci Rep. 2023 May 20;6(5):e1276. doi: 10.1002/hsr2.1276. eCollection 2023 May. Health Sci Rep. 2023. PMID: 37216052 Free PMC article.
-
Investigating a putative transcriptional regulatory protein encoded by Rv1719 gene of Mycobacterium tuberculosis.Protein J. 2022 Jun;41(3):424-433. doi: 10.1007/s10930-022-10062-9. Epub 2022 Jun 17. Protein J. 2022. PMID: 35715720
References
-
- Wayne LG. Dynamics of submerged growth of Mycobacterium tuberculosis under aerobic and microaerophilic conditions. Am Rev Respir Dis. 1976;114(4):807–811. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

