The RNA export factor Mex67 functions as a mobile nucleoporin
- PMID: 31753862
- PMCID: PMC6891080
- DOI: 10.1083/jcb.201909028
The RNA export factor Mex67 functions as a mobile nucleoporin
Abstract
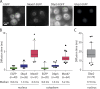
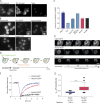
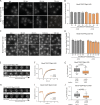
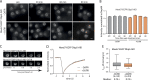
The RNA export factor Mex67 is essential for the transport of mRNA through the nuclear pore complex (NPC) in yeast, but the molecular mechanism of this export process remains poorly understood. Here, we use quantitative fluorescence microscopy techniques in live budding yeast cells to investigate how Mex67 facilitates mRNA export. We show that Mex67 exhibits little interaction with mRNA in the nucleus and localizes to the NPC independently of mRNA, occupying a set of binding sites offered by FG repeats in the NPC. The ATPase Dbp5, which is thought to remove Mex67 from transcripts, does not affect the interaction of Mex67 with the NPC. Strikingly, we find that the essential function of Mex67 is spatially restricted to the NPC since a fusion of Mex67 to the nucleoporin Nup116 rescues a deletion of MEX67 Thus, Mex67 functions as a mobile NPC component, which receives mRNA export substrates in the central channel of the NPC to facilitate their translocation to the cytoplasm.
© 2019 Derrer et al.
Figures

References
-
- Ben-Yishay R., Mor A., Shraga A., Ashkenazy-Titelman A., Kinor N., Schwed-Gross A., Jacob A., Kozer N., Kumar P., Garini Y., and Shav-Tal Y.. 2019. Imaging within single NPCs reveals NXF1's role in mRNA export on the cytoplasmic side of the pore. J. Cell Biol. 218:2962–2981. 10.1083/jcb.201901127 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

