Personalised analytics for rare disease diagnostics
- PMID: 31754101
- PMCID: PMC6872807
- DOI: 10.1038/s41467-019-13345-5
Personalised analytics for rare disease diagnostics
Abstract
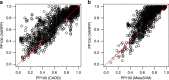
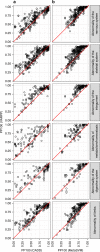
Whole genome and exome sequencing is a standard tool for the diagnosis of patients suffering from rare and other genetic disorders. The interpretation of the tens of thousands of variants returned from such tests remains a major challenge. Here we focus on the problem of prioritising variants with respect to the observed disease phenotype. We hypothesise that linking patterns of gene expression across multiple tissues to the phenotypes will aid in discovering disease causing variants. To test this, we construct classifiers that learn associations between tissue-specific gene expression and disease phenotypes. We find that using Genotype-Tissue Expression project (GTEx) expression data in conjunction with disease agnostic variant prioritisation methods (CADD or MetaSVM) results in consistent improvements in classification accuracy. Our method represents a previously overlooked avenue of utilising existing expression data for clinical diagnostics, and also opens the door to use of other functional genomic data sets in the same manner.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

