Coordination of cohabiting phage elements supports bacteria-phage cooperation
- PMID: 31754112
- PMCID: PMC6872733
- DOI: 10.1038/s41467-019-13296-x
Coordination of cohabiting phage elements supports bacteria-phage cooperation
Abstract
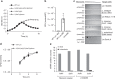
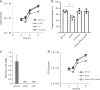
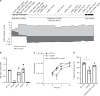
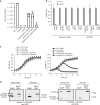
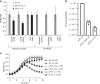
Bacterial pathogens often carry multiple prophages and other phage-derived elements within their genome, some of which can produce viral particles in response to stress. Listeria monocytogenes 10403S harbors two phage elements in its chromosome, both of which can trigger bacterial lysis under stress: an active prophage (ϕ10403S) that promotes the virulence of its host and can produce infective virions, and a locus encoding phage tail-like bacteriocins. Here, we show that the two phage elements are co-regulated, with the bacteriocin locus controlling the induction of the prophage and thus its activity as a virulence-associated molecular switch. More specifically, a metalloprotease encoded in the bacteriocin locus is upregulated in response to stress and acts as an anti-repressor for CI-like repressors encoded in each phage element. Our results provide molecular insight into the phenomenon of polylysogeny and its intricate adaptation to complex environments.
Conflict of interest statement
The authors declare no competing interests.
Figures
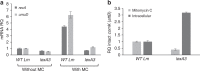

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

