Dissection of major cancer gene variants in subsets of circulating tumor cells in advanced breast cancer
- PMID: 31754145
- PMCID: PMC6872745
- DOI: 10.1038/s41598-019-53660-x
Dissection of major cancer gene variants in subsets of circulating tumor cells in advanced breast cancer
Abstract
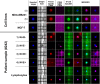
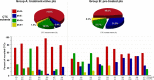
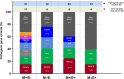
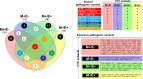
Enumeration of circulating tumor cells (CTCs) may reflect the metastatic potential of breast cancer (BC). By using the DEPArray, we investigated CTCs with respect to their epithelial-to-mesenchymal transition phenotype and compared their genomic heterogeneity with tissue biopsies. Seventeen stage IV BC patients were enrolled. Pre-enriched CTC suspensions were stained with fluorescent-labeled antibodies to epithelial (E) and mesenchymal (M) markers. CTC samples were processed by DEPArray system and clustered in relation to their markers. DNA from CTCs, as well as from primary tumor samples, was sequenced by next generation sequencing to assess the mutational state of 50 major cancer-related genes. We identified four different CTC subsets that harbored different gene variants. The most heterogenous CTC subsets included the M+/E- phenotype, which, however, expressed only 7 repeatedly mutated genes, while in the M-/E+ subset multiple mutations affected only 2 out of 50 genes. When matching all gene variants among CTC subsets, a small number of mutations was shared by only 4 genes, namely ATM, FGFR3, PIK3CA, and TP53 that, however, were absent in primary tumors. Our results postulate that the detected mutations in all CTC subsets may be considered as genomic markers of metastatic dissemination to be investigated during early stages of BC.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Mutational analysis of single circulating tumor cells by next generation sequencing in metastatic breast cancer.Oncotarget. 2016 May 3;7(18):26107-19. doi: 10.18632/oncotarget.8431. Oncotarget. 2016. PMID: 27034166 Free PMC article.
-
Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients.Mol Oncol. 2015 Apr;9(4):749-57. doi: 10.1016/j.molonc.2014.12.001. Epub 2014 Dec 9. Mol Oncol. 2015. PMID: 25539732 Free PMC article.
-
Mutational studies on single circulating tumor cells isolated from the blood of inflammatory breast cancer patients.Breast Cancer Res Treat. 2017 Jun;163(2):219-230. doi: 10.1007/s10549-017-4176-x. Epub 2017 Mar 7. Breast Cancer Res Treat. 2017. PMID: 28271309 Free PMC article.
-
Progress and challenges of sequencing and analyzing circulating tumor cells.Cell Biol Toxicol. 2018 Oct;34(5):405-415. doi: 10.1007/s10565-017-9418-5. Epub 2017 Nov 22. Cell Biol Toxicol. 2018. PMID: 29168077 Free PMC article. Review.
-
Dissecting the Heterogeneity of Circulating Tumor Cells in Metastatic Breast Cancer: Going Far Beyond the Needle in the Haystack.Int J Mol Sci. 2016 Oct 24;17(10):1775. doi: 10.3390/ijms17101775. Int J Mol Sci. 2016. PMID: 27783057 Free PMC article. Review.
Cited by
-
The Polemic Diagnostic Role of TP53 Mutations in Liquid Biopsies from Breast, Colon and Lung Cancers.Cancers (Basel). 2020 Nov 12;12(11):3343. doi: 10.3390/cancers12113343. Cancers (Basel). 2020. PMID: 33198130 Free PMC article. Review.
-
The Role and Therapeutic Targeting of CCR5 in Breast Cancer.Cells. 2023 Sep 8;12(18):2237. doi: 10.3390/cells12182237. Cells. 2023. PMID: 37759462 Free PMC article. Review.
-
Liquid Biopsy in Colorectal Carcinoma: Clinical Applications and Challenges.Cancers (Basel). 2020 May 27;12(6):1376. doi: 10.3390/cancers12061376. Cancers (Basel). 2020. PMID: 32471160 Free PMC article. Review.
-
Improving the Prognostic and Predictive Value of Circulating Tumor Cell Enumeration: Is Longitudinal Monitoring the Answer?Int J Mol Sci. 2024 Oct 2;25(19):10612. doi: 10.3390/ijms251910612. Int J Mol Sci. 2024. PMID: 39408942 Free PMC article. Review.
-
Dissecting Molecular Heterogeneity of Circulating Tumor Cells (CTCs) from Metastatic Breast Cancer Patients through Copy Number Aberration (CNA) and Single Nucleotide Variant (SNV) Single Cell Analysis.Cancers (Basel). 2022 Aug 14;14(16):3925. doi: 10.3390/cancers14163925. Cancers (Basel). 2022. PMID: 36010918 Free PMC article.
References
-
- Hayes DF, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:4218–4224. doi: 10.1158/1078-0432.CCR-05-2821. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

