Effects of sampling strategy and DNA extraction on human skin microbiome investigations
- PMID: 31754146
- PMCID: PMC6872721
- DOI: 10.1038/s41598-019-53599-z
Effects of sampling strategy and DNA extraction on human skin microbiome investigations
Abstract
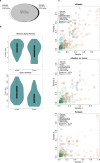
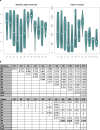
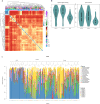
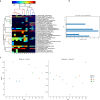
The human skin is colonized by a wide array of microorganisms playing a role in skin disorders. Studying the skin microbiome provides unique obstacles such as low microbial biomass. The objective of this study was to establish methodology for skin microbiome analyses, focusing on sampling technique and DNA extraction. Skin swabs and scrapes were collected from 9 healthy adult subjects, and DNA extracted using 12 commercial kits. All 165 samples were sequenced using the 16S rRNA gene. Comparing the populations captured by eSwabs and scrapes, 99.3% of sequences overlapped. Using eSwabs yielded higher consistency. The success rate of library preparation applying different DNA extraction kits ranged from 39% to 100%. Some kits had higher Shannon alpha-diversity. Metagenomic shotgun analyses were performed on a subset of samples (N = 12). These data indicate that a reduction of human DNA from 90% to 57% is feasible without lowering the success of 16S rRNA library preparation and without introducing taxonomic bias. Using swabs is a reliable technique to investigate the skin microbiome. DNA extraction methodology is crucial for success of sequencing and adds a substantial amount of variation in microbiome analyses. Reduction of host DNA is recommended for interventional studies applying metagenomics.
Conflict of interest statement
All authors declare that they have no financial competing interests. J.D.J. is head of the cosmetic counsel (unpaid position) advisory to the Danish Environmental Protection Agency.
Figures
References
-
- Altunbulakli Can, Reiger Matthias, Neumann Avidan U., Garzorz-Stark Natalie, Fleming Megan, Huelpuesch Claudia, Castro-Giner Francesc, Eyerich Kilian, Akdis Cezmi A., Traidl-Hoffmann Claudia. Relations between epidermal barrier dysregulation and Staphylococcus species–dominated microbiome dysbiosis in patients with atopic dermatitis. Journal of Allergy and Clinical Immunology. 2018;142(5):1643-1647.e12. doi: 10.1016/j.jaci.2018.07.005. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources

