Helicobacter pylori in ancient human remains
- PMID: 31754290
- PMCID: PMC6861846
- DOI: 10.3748/wjg.v25.i42.6289
Helicobacter pylori in ancient human remains
Abstract
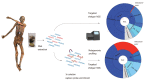
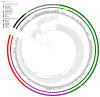
The bacterium Helicobacter pylori (H. pylori) infects the stomachs of approximately 50% of all humans. With its universal occurrence, high infectivity and virulence properties it is considered as one of the most severe global burdens of modern humankind. It has accompanied humans for many thousands of years, and due to its high genetic variability and vertical transmission, its population genetics reflects the history of human migrations. However, especially complex demographic events such as the colonisation of Europe cannot be resolved with population genetic analysis of modern H. pylori strains alone. This is best exemplified with the reconstruction of the 5300-year-old H. pylori genome of the Iceman, a European Copper Age mummy. Our analysis provided precious insights into the ancestry and evolution of the pathogen and underlined the high complexity of ancient European population history. In this review we will provide an overview on the molecular analysis of H. pylori in mummified human remains that were done so far and we will outline methodological advancements in the field of ancient DNA research that support the reconstruction and authentication of ancient H. pylori genome sequences.
Keywords: Helicobacter pylori; Ancient DNA; Ancient gut contents; Coprolites; Evolution; Iceman.
©The Author(s) 2019. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: No potential conflicts of interest.
Figures


References
-
- Malfertheiner P, Chan FK, McColl KE. Peptic ulcer disease. Lancet. 2009;374:1449–1461. - PubMed
-
- Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI, Yamaoka Y, Mégraud F, Otto K, Reichard U, Katzowitsch E, Wang X, Achtman M, Suerbaum S. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. - PubMed
-
- Moodley Y, Linz B. Helicobacter pylori Sequences Reflect Past Human Migrations. Genome Dyn. 2009;6:62–74. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

