A guide to computational methods for G-quadruplex prediction
- PMID: 31754698
- PMCID: PMC6943126
- DOI: 10.1093/nar/gkz1097
A guide to computational methods for G-quadruplex prediction
Erratum in
-
A guide to computational methods for G-quadruplex prediction.Nucleic Acids Res. 2020 Feb 20;48(3):1603. doi: 10.1093/nar/gkaa033. Nucleic Acids Res. 2020. PMID: 31943112 Free PMC article. No abstract available.
Abstract
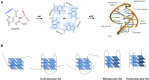
Guanine-rich nucleic acids can fold into the non-B DNA or RNA structures called G-quadruplexes (G4). Recent methodological developments have allowed the characterization of specific G-quadruplex structures in vitro as well as in vivo, and at a much higher throughput, in silico, which has greatly expanded our understanding of G4-associated functions. Typically, the consensus motif G3+N1-7G3+N1-7G3+N1-7G3+ has been used to identify potential G-quadruplexes from primary sequence. Since, various algorithms have been developed to predict the potential formation of quadruplexes directly from DNA or RNA sequences and the number of studies reporting genome-wide G4 exploration across species has rapidly increased. More recently, new methodologies have also appeared, proposing other estimates which consider non-canonical sequences and/or structure propensity and stability. The present review aims at providing an updated overview of the current open-source G-quadruplex prediction algorithms and straightforward examples of their implementation.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Sen D., Gilbert W.. Formation of parallel four-stranded complexes by guanine rich motifs in DNA and its implications for meiosis. Nature. 1988; 334:364–366. - PubMed
-
- Sen D., Gilbert W.. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990; 334:410–414. - PubMed
-
- Simonsson T. G-quadruplex DNA structures–variations on a theme. Biol. Chem. 2001; 382:621–628. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

