Poly(ADP-ribose) polymerase enzymes and the maintenance of genome integrity
- PMID: 31754726
- PMCID: PMC11104942
- DOI: 10.1007/s00018-019-03366-0
Poly(ADP-ribose) polymerase enzymes and the maintenance of genome integrity
Abstract
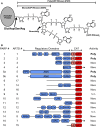
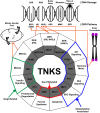
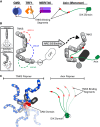
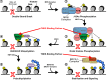
DNA damage response (DDR) relies on swift and accurate signaling to rapidly identify DNA lesions and initiate repair. A critical DDR signaling and regulatory molecule is the posttranslational modification poly(ADP-ribose) (PAR). PAR is synthesized by a family of structurally and functionally diverse proteins called poly(ADP-ribose) polymerases (PARPs). Although PARPs share a conserved catalytic domain, unique regulatory domains of individual family members endow PARPs with unique properties and cellular functions. Family members PARP-1, PARP-2, and PARP-3 (DDR-PARPs) are catalytically activated in the presence of damaged DNA and act as damage sensors. Family members tankyrase-1 and closely related tankyrase-2 possess SAM and ankyrin repeat domains that regulate their diverse cellular functions. Recent studies have shown that the tankyrases share some overlapping functions with the DDR-PARPs, and even perform novel functions that help preserve genomic integrity. In this review, we briefly touch on DDR-PARP functions, and focus on the emerging roles of tankyrases in genome maintenance. Preservation of genomic integrity thus appears to be a common function of several PARP family members, depicting PAR as a multifaceted guardian of the genome.
Keywords: DDR, DNA damage response; PAR, poly(ADP-ribose); PARP, poly(ADP-ribose) polymerase; Tankyrase.
Figures

References
-
- Hoeijmakers JHJ. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

