NeoFuse: predicting fusion neoantigens from RNA sequencing data
- PMID: 31755900
- PMCID: PMC7141848
- DOI: 10.1093/bioinformatics/btz879
NeoFuse: predicting fusion neoantigens from RNA sequencing data
Abstract
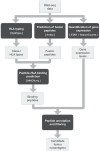
Summary: Gene fusions can generate immunogenic neoantigens that mediate anticancer immune responses. However, their computational prediction from RNA sequencing (RNA-seq) data requires deep bioinformatics expertise to assembly a computational workflow covering the prediction of: fusion transcripts, their translated proteins and peptides, Human Leukocyte Antigen (HLA) types, and peptide-HLA binding affinity. Here, we present NeoFuse, a computational pipeline for the prediction of fusion neoantigens from tumor RNA-seq data. NeoFuse can be applied to cancer patients' RNA-seq data to identify fusion neoantigens that might expand the repertoire of suitable targets for immunotherapy.
Availability and implementation: NeoFuse source code and documentation are available under GPLv3 license at https://icbi.i-med.ac.at/NeoFuse/.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2020. Published by Oxford University Press.
Figures
References
-
- Finotello F. et al. (2019) Next- generation computational tools for interrogating cancer immunity. Nat. Rev. Genet., 20, 724. - PubMed
-
- Haas B. et al. (2017) STAR-fusion: fast and accurate fusion transcript detection from RNA-seq. BioRxiv, 120295. doi: 10.1101/120295.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials