Integrated analysis of miRNA landscape and cellular networking pathways in stage-specific prostate cancer
- PMID: 31756185
- PMCID: PMC6874298
- DOI: 10.1371/journal.pone.0224071
Integrated analysis of miRNA landscape and cellular networking pathways in stage-specific prostate cancer
Abstract
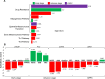
Dysregulation of miRNAs has been demonstrated in several human malignancies including prostate cancer. Due to tissue limitation and variable disease progression, stage-specific miRNAs changes in prostate cancer is unknown. Using chip-based microarray, we investigated global miRNA expression in human prostate cancer LNCaP, PC3, DU145 and 22Rv1 cells representing early-stage, advanced-stage and castration resistant prostate cancer in comparison with normal prostate epithelial cells. A total of 292 miRNAs were differentially expressed with 125 upregulated and 167 downregulated. These miRNAs were involved in pathways including drug resistance drug-efflux, adipogenesis, epithelial-to-mesenchymal transition, bone metamorphosis, and Th1/Th2 signaling. Regulation of miRNAs were interlinked with upstream regulators such as Argonaut 2 (AGO2), Double-Stranded RNA-Specific Endoribonuclease (DICER1), Sjogren syndrome antigen B (SSB), neurofibromatosis 2 (NF2), and peroxisome proliferator activated receptor alpha (PPARA), activated during stage-specific disease progression. Candidate target genes and pathways dysregulated in stage-specific prostate cancer were identified using CS-miRTar database and confirmed in clinical specimens. Integrative network analysis suggested some genes targeted by miRNAs include miR-17, let7g, miR-146, miR-204, miR-205, miR-221, miR-301 and miR-520 having a major effect on their dysregulation in prostate cancer. MiRNA-microarray analysis further identified miR-130a, miR-181, miR-328, miR146 and miR-200 as a panel of novel miRNAs associated with drug resistance drug-efflux and epithelial-to-mesenchymal transition in prostate cancer. Our findings provide evidence on miRNA dysregulation and its association with key functional components in stage-specific prostate cancer.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

