Genes with human-specific features are primarily involved with brain, immune and metabolic evolution
- PMID: 31757203
- PMCID: PMC6873653
- DOI: 10.1186/s12859-019-2886-2
Genes with human-specific features are primarily involved with brain, immune and metabolic evolution
Abstract
Background: Humans have adapted to widespread changes during the past 2 million years in both environmental and lifestyle factors. This is evident in overall body alterations such as average height and brain size. Although we can appreciate the uniqueness of our species in many aspects, molecular variations that drive such changes are far from being fully known and explained. Comparative genomics is able to determine variations in genomic sequence that may provide functional information to better understand species-specific adaptations. A large number of human-specific genomic variations have been reported but no currently available dataset comprises all of these, a problem which contributes to hinder progress in the field.
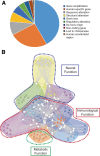
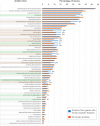
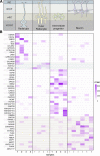
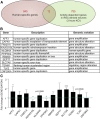
Results: Here we critically update high confidence human-specific genomic variants that mostly associate with protein-coding regions and find 856 related genes. Events that create such human-specificity are mainly gene duplications, the emergence of novel gene regions and sequence and structural alterations. Functional analysis of these human-specific genes identifies adaptations to brain, immune and metabolic systems to be highly involved. We further show that many of these genes may be functionally associated with neural activity and generating the expanded human cortex in dynamic spatial and temporal contexts.
Conclusions: This comprehensive study contributes to the current knowledge by considerably updating the number of human-specific genes following a critical bibliographic survey. Human-specific genes were functionally assessed for the first time to such extent, thus providing unique information. Our results are consistent with environmental changes, such as immune challenges and alterations in diet, as well as neural sophistication, as significant contributors to recent human evolution.
Keywords: Brain; Gene expression; Glia; Human-specific; Metabolism; Neuron.
Conflict of interest statement
All authors declare that they have no competing interests.
Figures
References
-
- Tattersall I. Why was human evolution so rapid? In:Human Paleontology and Prehistory. 2017:1:1–9.
-
- Zink KD, Lieberman DE. Impact of meat and lower Palaeolithic food processing techniques on chewing in humans. Nature. 2017;531:500–503. - PubMed
-
- Weyrich LS, Duchene S, Soubrier J, Arriola L, Llamas B, Breen J, Morris AG, Alt KW, Caramelli D, Dresely V, et al. Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature. 2017;544:357–361. - PubMed
-
- Dutrillaux B. Chromosomal evolution in primates: tentative phylogeny from Microcebus murinus (prosimian) to man. Hum Genet. 1979;48:251–314. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

