The Anti-Aggregation Holdase Hsp33 Promotes the Formation of Folded Protein Structures
- PMID: 31757359
- PMCID: PMC6953636
- DOI: 10.1016/j.bpj.2019.10.040
The Anti-Aggregation Holdase Hsp33 Promotes the Formation of Folded Protein Structures
Abstract
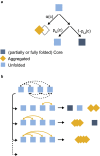
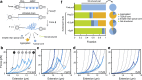
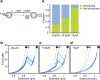
Holdase chaperones are known to be central to suppressing aggregation, but how they affect substrate conformations remains poorly understood. Here, we use optical tweezers to study how the holdase Hsp33 alters folding transitions within single maltose binding proteins and aggregation transitions between maltose binding protein substrates. Surprisingly, we find that Hsp33 not only suppresses aggregation but also guides the folding process. Two modes of action underlie these effects. First, Hsp33 binds unfolded chains, which suppresses aggregation between substrates and folding transitions within substrates. Second, Hsp33 binding promotes substrate states in which most of the chain is folded and modifies their structure, possibly by intercalating its intrinsically disordered regions. A statistical ensemble model shows how Hsp33 function results from the competition between these two contrasting effects. Our findings reveal an unexpectedly comprehensive functional repertoire for Hsp33 that may be more prevalent among holdases and dispels the notion of a strict chaperone hierarchy.
Copyright © 2019. Published by Elsevier Inc.
Figures
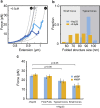
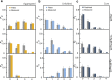
References
-
- Buchberger A., Bukau B., Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol. Cell. 2010;40:238–252. - PubMed
-
- Skowyra D., Georgopoulos C., Zylicz M. The E. coli dnaK gene product, the hsp70 homolog, can reactivate heat-inactivated RNA polymerase in an ATP hydrolysis-dependent manner. Cell. 1990;62:939–944. - PubMed
-
- Sharma S., Chakraborty K., Hartl F.U. Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell. 2008;133:142–153. - PubMed
-
- Thirumalai D., Lorimer G.H. Chaperonin-mediated protein folding. Annu. Rev. Biophys. Biomol. Struct. 2001;30:245–269. - PubMed
-
- Haslbeck M., Franzmann T., Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 2005;12:842–846. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

