Functional analysis of genetic variants in the high-risk breast cancer susceptibility gene PALB2
- PMID: 31757951
- PMCID: PMC6876638
- DOI: 10.1038/s41467-019-13194-2
Functional analysis of genetic variants in the high-risk breast cancer susceptibility gene PALB2
Abstract
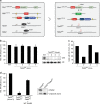
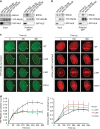
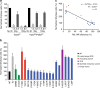
Heterozygous carriers of germ-line loss-of-function variants in the DNA repair gene PALB2 are at a highly increased lifetime risk for developing breast cancer. While truncating variants in PALB2 are known to increase cancer risk, the interpretation of missense variants of uncertain significance (VUS) is in its infancy. Here we describe the development of a relatively fast and easy cDNA-based system for the semi high-throughput functional analysis of 48 VUS in human PALB2. By assessing the ability of PALB2 VUS to rescue the DNA repair and checkpoint defects in Palb2 knockout mouse embryonic stem (mES) cells, we identify various VUS in PALB2 that impair its function. Three VUS in the coiled-coil domain of PALB2 abrogate the interaction with BRCA1, whereas several VUS in the WD40 domain dramatically reduce protein stability. Thus, our functional assays identify damaging VUS in PALB2 that may increase cancer risk.
Conflict of interest statement
The authors declare no competing interests.
Figures




Comment in
-
PALB2 Genetic Variants: Can Functional Assays Assist Translation?Trends Cancer. 2020 Apr;6(4):263-265. doi: 10.1016/j.trecan.2020.01.017. Epub 2020 Feb 13. Trends Cancer. 2020. PMID: 32209438
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

