Disentangling the role of Africa in the global spread of H5 highly pathogenic avian influenza
- PMID: 31757953
- PMCID: PMC6874648
- DOI: 10.1038/s41467-019-13287-y
Disentangling the role of Africa in the global spread of H5 highly pathogenic avian influenza
Abstract
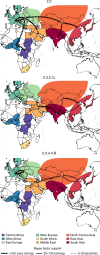
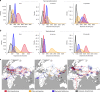
The role of Africa in the dynamics of the global spread of a zoonotic and economically-important virus, such as the highly pathogenic avian influenza (HPAI) H5Nx of the Gs/GD lineage, remains unexplored. Here we characterise the spatiotemporal patterns of virus diffusion during three HPAI H5Nx intercontinental epidemic waves and demonstrate that Africa mainly acted as an ecological sink of the HPAI H5Nx viruses. A joint analysis of host dynamics and continuous spatial diffusion indicates that poultry trade as well as wild bird migrations have contributed to the virus spreading into Africa, with West Africa acting as a crucial hotspot for virus introduction and dissemination into the continent. We demonstrate varying paths of avian influenza incursions into Africa as well as virus spread within Africa over time, which reveal that virus expansion is a complex phenomenon, shaped by an intricate interplay between avian host ecology, virus characteristics and environmental variables.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- WHO | Cumulative number of confirmed human cases of avian influenza A(H5N1) reported to WHO. WHO (2019). Available at: https://www.who.int/influenza/human_animal_interface/2019_04_09_tableH5N.... Accessed April 20, 2019.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

