Multi-site reproducibility of a human immunophenotyping assay in whole blood and peripheral blood mononuclear cells preparations using CyTOF technology coupled with Maxpar Pathsetter, an automated data analysis system
- PMID: 31758746
- PMCID: PMC7543682
- DOI: 10.1002/cyto.b.21858
Multi-site reproducibility of a human immunophenotyping assay in whole blood and peripheral blood mononuclear cells preparations using CyTOF technology coupled with Maxpar Pathsetter, an automated data analysis system
Abstract
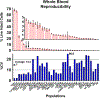
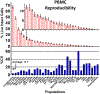
High-dimensional mass cytometry data potentially enable a comprehensive characterization of immune cells. In order to positively affect clinical trials and translational clinical research, this advanced technology needs to demonstrate a high reproducibility of results across multiple sites for both peripheral blood mononuclear cells (PBMC) and whole blood preparations. A dry 30-marker broad immunophenotyping panel and customized automated analysis software were recently engineered and are commercially available as the Fluidigm® Maxpar® Direct™ Immune Profiling Assay™. In this study, seven sites received whole blood and six sites received PBMC samples from single donors over a 2-week interval. Each site labeled replicate samples and acquired data on Helios™ instruments using an assay-specific acquisition template. All acquired sample files were then automatically analyzed by Maxpar Pathsetter™ software. A cleanup step eliminated debris, dead cells, aggregates, and normalization beads. The second step automatically enumerated 37 immune cell populations and performed label intensity assessments on all 30 markers. The inter-site reproducibility of the 37 quantified cell populations had consistent population frequencies, with an average %CV of 14.4% for whole blood and 17.7% for PBMC. The dry reagent coupled with automated data analysis is not only convenient but also provides a high degree of reproducibility within and among multiple test sites resulting in a comprehensive yet practical solution for deep immune phenotyping.
Keywords: cytometry automation; cytometry standardization; kits; percentage precision.
© 2019 International Clinical Cytometry Society.
Conflict of interest statement
CONFLICT OF INTEREST
Authors Bagwell, Hunsberger, Hill, Herbert, Bray, Selvanantham, Li, Inokuma, Goldberger, and Stelzer are currently employed or were employed by either Verity Software House or Fluidigm Corporation. Author Inokuma is a consultant for both Verity Software House and Fluidigm Corporation. This manuscript describes a component of the product, Fluidigm Maxpar Pathsetter, which was a collaborative effort between these two companies.
Figures

References
-
- Bagwell C. (2010). Probability state modeling: A new paradigm for cytometric analysis In Litwin V. & Marder P. (Eds.), Flow cytometry in drug discovery and development (p. 281). Hoboken, NJ: John Wiley and Sons Inc.
-
- Bagwell CB (2018). Chapter 2: High-dimensional modeling for cytometry: Building rock solid models using GemStonetm™ and Verity Cen-se′™ high-dimensional t-SNE mapping In Hawley TS & Hawley RG (Eds.), Methods in molecular biology (Vol. 1678, p. 2018). New York: Springer Science+Business Media LLC. - PubMed
-
- Bagwell CB, Hunsberger BC, Herbert DJ, Munson ME, Hill BL, Bray CM, & Preffer FI (2015). Probability state modeling theory. Cytometry Part A, 87, 646–660. - PubMed
-
- Bagwell CB, Leipold M, Maecker H, & Stelzer G. (2016). Highdimensional modeling of peripheral blood mononuclear cells from a Helios Instrument. Seattle, Washington: Washington State Convention Center.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

